Non-small cell lung cancer (NSCLC) accounts for approximately 80–85% of lung cancer cases.[1,2] In recent years, the diagnostic strategies and treatment methods of NSCLC have been continuously developed, but patients showed unfavorable prognoses, with low 5-year survival rates.[3,4] Hence, further exploring functional genes related to the onset and advance of NSCLC is beneficial for NSCLC therapy.
Sulfatase 1 (SULF1) is one of extracellular sulfatases and functions in signal transduction, cell development, tumorigenesis, muscle regeneration, and immune regulation.[5] A study has shown that SULF1 is augmented in cervical cancer tissues and is prominently linked to its poor prognosis.[6] Moreover, a research has pointed out that SULF1 is carcinogenic in multiple malignant tumors and linked to poor survival,[7] but the role of SULF1 in NSCLC remains unclear.
As a member of the receptor tyrosine kinase ErbB family, epidermal growth factor receptor (EGFR) has a crucial function in physiological events. It usually mutates or is overexpressed in different cancer types and is a therapeutic target for a variety of clinical cancer.[8,9] Osimertinib is an EGFR–tyrosine kinase inhibitor and the standard therapy for patients with advanced NSCLC concomitant with EGFR mutation; its safety and efficacy have been confirmed.[10] EGFR exerts physiological functions through ligand binding. After combination, tyrosine residues in EGFR undergo dimerization and autophosphorylation, and EGFR activates downstream signaling through mitogen-activated protein kinase ( MAPK) and phosphatidylinositol-3-kinase (PI3K) pathways, thereby regulating physiologic properties in tumor cells.[11] The EGFR/MAPK signaling pathway is a potential therapeutic target against NSCLC because of its contribution to cancer cell proliferation, angiogenesis, and metastasis.[12,13] Long non-coding ribonucleic acid nuclear-enriched abundant transcript 1 knockdown can inhibit MAPK signaling by modulating SULF1 in NSCLC cells.[14] However, the effects of SULF1 on the EGFR/MAPK signaling pathway and whether it is involved in the course of NSCLC have not been reported. Our research objective was to examine the effects and potential molecular mechanisms of SULF1 in NSCLC. The SULF1 expression in the tumor tissues was assessed, and the changes affected by SULF1 knockdown in NSCLC were evaluated in vitro. This study may assist in the exploration of potential therapeutic targets for NSCLC.
MATERIAL AND METHODS Cell cultivation and small interfering RNA (siRNA) transfectionAmerican Tissue Culture Collection (Manassas, VA, USA) provided the NSCLC cell lines: NCI-H1299 (CRL-5803) and HCC827 (CRL-2868) cells. Human normal lung epithelial cells BEAS-2B (TCH-C132), human normal lung epithelial cells, was obtained from Suzhou Haixing Biotechnology Co., Ltd. (Suzhou, China). Short tandem repeat profiling and mycoplasma testing were carried out before experiments. Dulbecco’s modified Eagle’s medium (11885084) containing 10% fetal bovine serum (10099–141) and penicillin/streptomycin (15070063) were used for cell cultivation. Reagents for cell cultivation were sourced from Gibco (Thermo Fisher Scientific, Waltham, MA, USA). The temperature and carbon dioxide(CO2) supply of the incubator was set at 37°C and 5%, respectively.
siRNA was designed and synthesized by Guangzhou Ruibo Biotechnology Co., Ltd. (Guangzhou, China), and sequences were as follows: Negative control (si-NC; 5'-AUGCUGATCAGUGUCGATU-3') and siRNA SULF1 (si-SULF1; 5'-CGGGAAGTATGTGCACAATCACA-3'). Lipofectamine 3000 (L3000015, Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used in transfecting NSCLC cells with si-NC or si-SULF1. After 48 h of transfection, subsequent experiments were undertaken using the collected cells. NSC228155 (100 μM, HY-101084, MCE, Shanghai, China), an EGFR/MAPK pathway agonist, was used for processing NCI-H1299 cells.
Quantitative real-time polymerase chain reaction (qRT-PCR)Trizol (Sigma-Aldrich, T9424, Saint Louis, MO, USA) was used for total RNA extraction. Complementary deoxyribonucleic acid synthesis was conducted in accordance with the instructions of HiScript III RT SuperMix (R323-01, Vazyme Biotech Co., Ltd., Nanjing, China). A reaction system for fluorescence quantitative polymerase chain reaction (qPCR) detection was established using an AceQ qPCR Synergy Brands Green master kit (Vazyme Biotech Co. Ltd., Q111–02, Nanjing, China). The reaction procedures were as follows: 95°C for 3 min; 95°C 5 s, 56°C 10 s, 72°C 25 s; 40 cycles. After the reaction, the cycle threshold (Ct) value was obtained. The internal reference was glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the analytical approach was the 2−∆∆Ct method. The primer sequences were as follows: SULF1 forward: 5'-TGGCGAGAATGGCTTGGATTA-3', SULF1 reverse: 5'-TAACGGGCCTATGGGGATACA-3'; GAPDH forward: 5'-TTGAGGTCAATGAAGGGGTC-3', GAPDH reverse: 5'-GAAGGTGAAGGTCGGAGTCA-3'.
Cell Counting Kit-8 (CCK-8) assayCells were cultured in 96-well plates (with density of 3 × 103 cells per well) until they to be adherent. Next, the plates were collected, and CCK-8 solution (10 μL, BestBio, BB-4202, Shanghai, China) was added to each well. Four hours later, microplate reader (MR-96A, Mindray, Shenzhen, China) was for the detection of optical density value (450 nm).
5-Ethynyl-2-deoxyuridine (EdU) assayCells inoculated in 96-well plates (3 × 103 cells per well) were placed in an incubator for 24 h. Phosphate buffer saline (PBS) was rinsed 3 times, and 100 μL of 5-Ethynyl-2-deoxyuridine ( EdU) staining solution was added to each well. The cells were stained for 2 h under dark condition. Cell fixation, Apollo staining, 4',6-diamidino-2-phenylindole staining (C1005, Beyotime, Shanghai, China), and fluorescence microscopy (Ts2/Ts2-FL, Nikon, Tokyo, Japan) were performed in accordance with the description of the EdU cell proliferation detection kit (Guangzhou RiboBio Co., Ltd., C10310-1, Guangzhou, China). Three fields of view were randomly selected for photographing. Image J software (version 1.4, National Institutes of Health, Bethesda, MD, USA) was for quantitatively analyzing.
Transwell assay Migration assayThe cells were inoculated in Transwell chambers (Merck, CLS3399, Kenilworth, NJ, USA) at a density 1×105 cells/well. A complete culture medium was added to the lower 24-well plate. After 24 h of incubation, the medium was discarded. Cells were subjected to PBS washing, 4% formaldehyde fixation for 4 h, and crystal violet solution (Solarbio, C8470, Beijing, China) staining for 10 min. After the upper cells were wiped away, migrating cells were observed under a microscope (DMIL LED, Leica, Wetzlar, Germany). Three fields of view were randomly selected for cell counting.
Invasion assayBefore cell inoculation, Transwell chambers supplementing Matrigel (Shanghai Fusheng Industrial Co., Ltd., FS-79064, Shanghai, China) was maintained at 37°C for 30 min. The rest of the procedures were the same as those in the migration experiment.
Western blotRadioimmunoprecipitation assay lysis buffer (Merck, 20-188, Kenilworth, NJ, USA) was utilized for total protein extraction. A bicinchoninic acid kit (Merck, BCA1-1KT, Kenilworth, NJ, USA) was used for quantification. Protein (20 μg) was added to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%) for separation and displaced to polyvinylidene fluoride membranes (Merck, IPFL00010, Kenilworth, NJ, USA). Upon blocking with 5% skimmed milk powder (Solarbio, D8340, Beijing, China), primary antibodies were added to the membranes for overnight incubation at 4°C. The primary antibodies were as follows: E-cadherin (1:1000; A22333) and GAPDH (1:10,000; AC001) purchased from Abclonal (Wuhan, China); N-cadherin (1:1000; 4061), vimentin (1:1000; 3932), c-Jun N-terminal kinase (JNK, 1:1000; 9252), and phospho-JNK (p-JNK, 1:1000; 4671) from CST (Danvers, MA, USA); EGFR (1:5000; 30139-1-AP), phospho-EGFR (p-EGFR, 1:1000; 30277-1-AP), extracellular signal-regulated kinase (ERK; 1:5000; 51068-1-AP), phospho-ERK (p-ERK; 1:1000; 28733-1-AP), p-p38 MAPK (1:1000; 28796-1-AP), and p38 MAPK (1:1000; 51115-1-AP) from Proteintech (Wuhan, China). Then, the membranes were incubated with goat anti-rabbit immunoglobulin G secondary antibody (1:5000; AS014, Abclonal, Wuhan, China) for 1 h. An enhanced chemiluminescence kit (Thermo Fisher Scientific, WP20005, Waltham, MA, USA) was utilized for exposure development. A Bio-Tanon imaging system (Tanon Science and Technology Co., Ltd., Shanghai, China) was used for visualization, imaging, and quantification. GAPDH was used as the endogenous control, and Image J software was used for quantitative analysis (grayscale analysis).
Clinical samplesNSCLC tissues and paraneoplastic tissues were obtained from 54 patients with NSCLC who received surgical resection in Ningbo Ninth Hospital from February 2021 to January 2022. NSCLC was diagnosed with computed tomography scan and histopathological biopsy. No preoperative chemotherapy or radiotherapy was administered to a patient before surgery. After cold PBS rinses, the tissues were stored at −80°C. SULF1 expression in the clinical tissue samples was evaluated using qRT-PCR. The general information of patients was collected, including age, sex, pathological type, differentiation, tumor node metastasis (TNM) stage, and lymph node metastasis. The patients were followed up once a month, and their survival status was recorded. The median follow-up period was 29 months (7–38 months). The ethics committee of Ningbo Ninth Hospital Medical Health Group provided approval (approval number NNHM-20210106). All the procedures complied with the Helsinki Declaration and have obtained informed consent from patients.
Statistical analysisThe Statistical Package for the Social Sciences 22.0 software (IBM, Armonk, NY, USA) and Graphpad Prism 9.0 (Graphpad, San Diego, CA, USA) were used for statistical analysis and drawing, respectively. The data were presented as mean and standard deviation. Intergroup comparisons were performed using Student’s t-test (for two groups), and one-way analysis of variance was performed with Tukey’s post hoc test (for multiple groups). Chi-square test was used for evaluating difference in count data, which was denoted as n (%). On the basis of the median SULF1 expression value, the patients were included in the high SULF1 expression group (SULF1 expression above the median) and low SULF1 expression group (SULF1 expression below the median). Difference in overall survival between two groups was evaluated through Kaplan–Meier analysis. The criterion for significant variation was P < 0.05.
RESULTS Knockdown of SULF1 abated NSCLC cell viabilityThe Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) database indicated that lung adenocarcinoma (LUAD, which is a type of NSCLC) tumor tissues have higher SULF1 expression levels than normal tissues (P < 0.05), [Figure 1a]. In contrast to human normal lung epithelial cells BEAS-2B, NSCLC cells showed prominently augmented SULF1 expression levels (P < 0.001), [Figure 1b]. Next, si-SULF1 and its negative control were transfected into NCI-H1299 and HCC827 cells. The SULF1 levels in the NSCLC cells were abated after transfected with si-SULF1 relative to those of transfected with si-NC (P < 0.001), [Figure 1c and d], which were measured through qRT-PCR. This result indicated that transfection successfully knocked down the SULF1 expression. The results from CCK-8 assay showed that versus the cell viability of siNC group, the cell viability in si-SULF1 group was prominently abated (P < 0.05), [Figure 1e and f]. The EdU-positive cell rates of the NCI-H1299 and HCC827 cells in the si-SULF1 group were considerably lower than the EdU-positive cell rate of the si-NC group (P < 0.01), [Figure 1g-j]. The rates were measured with EdU assay. Overall, the knockdown of SULF1 inhibited the activity of the NSCLC cells.
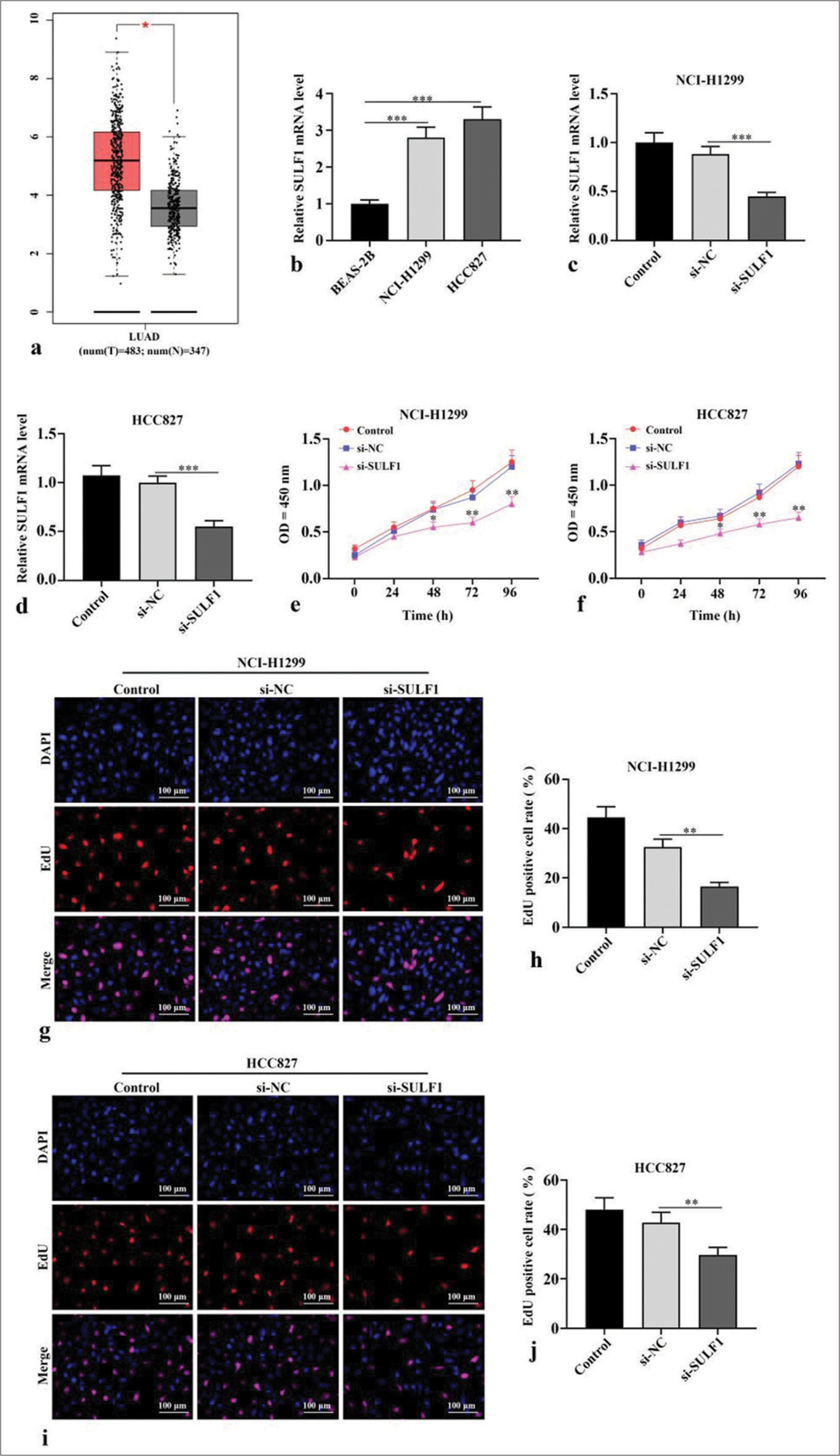
Export to PPT
Knockdown of SULF1 inhibits migration, invasion, and epithelial-to-mesenchymal transition (EMT) in the NSCLC cellsCompared with number of migration and invasion cells of the si-NC group, the number of migration and invasion cells in si-SULF1 group were prominently reduced (P < 0.01), [Figure 2a-f]. Epithelial-to-mesenchymal transition (EMT) -related proteins, including the epithelial cell marker E-cadherin and mesenchymal cell markers N-cadherin and vimentin, were evaluated through Western blot. Compared with the si-NC group, the si-SULF1 group exhibited prominently augmented E-cadherin protein level in the H1299 and HCC827 cells (P < 0.001) and abated N-cadherin and vimentin protein levels (P < 0.01), [Figure 2g-i]. Confirming that SULF1 knockdown repressed the malignant biological behavior of the NSCLC cells, including migration, invasion, and EMT.
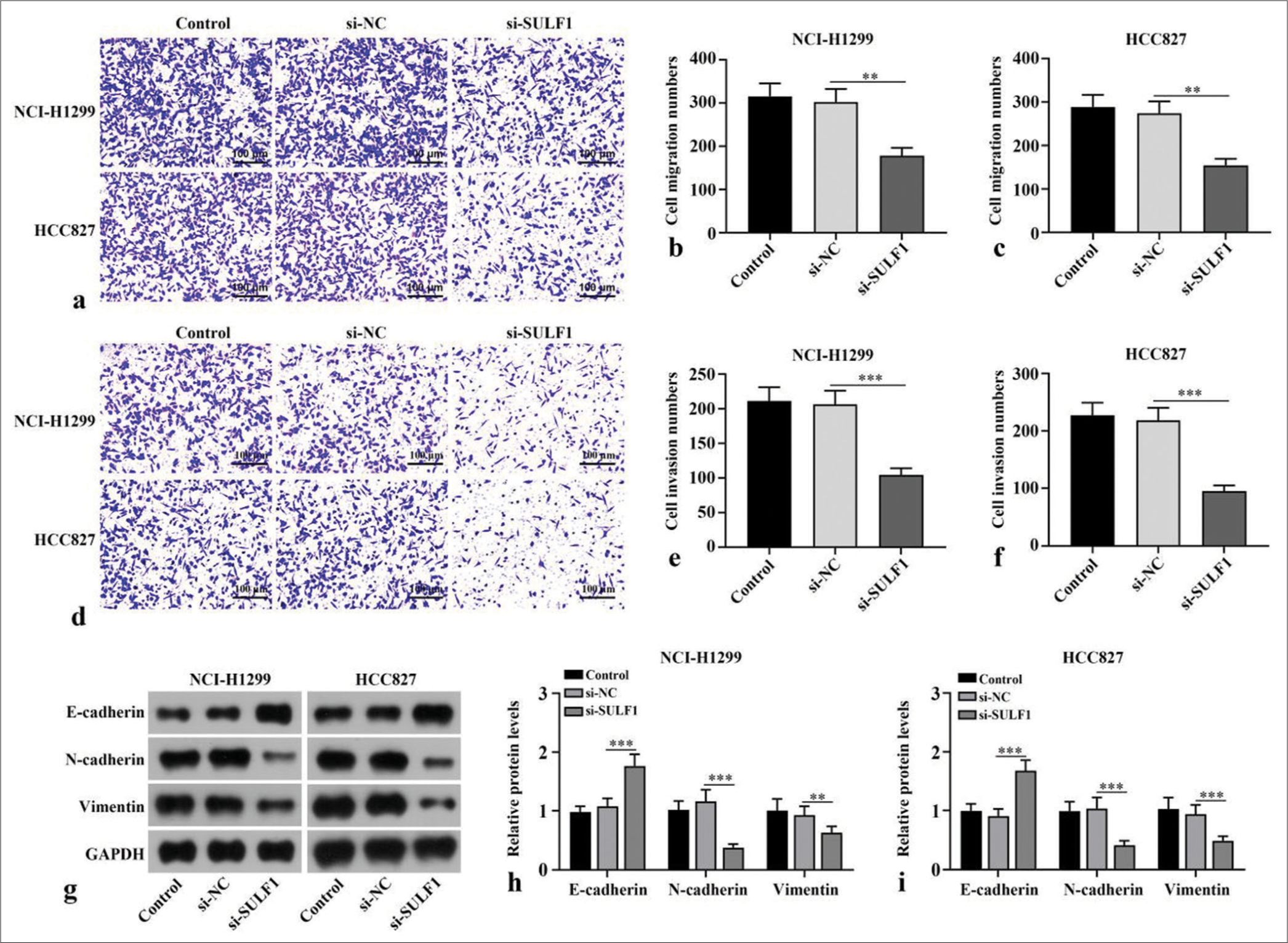
Export to PPT
Knockdown of SULF1 abated EGFR/MAPK signaling pathwayp-EGFR/EGFR, p-p38 MAPK/p38 MAPK, p-ERK/ERK, and p-JNK/JNK ratios in the NCI-H1299 and HCC827 cells transfected with si-SULF1 were prominently abated relative to those in the si-NC group (P < 0.001), [Figure 3a-j], as indicated by the Western blot results. SULF1 knockdown can abate the EGFR/MAPK signaling pathway.
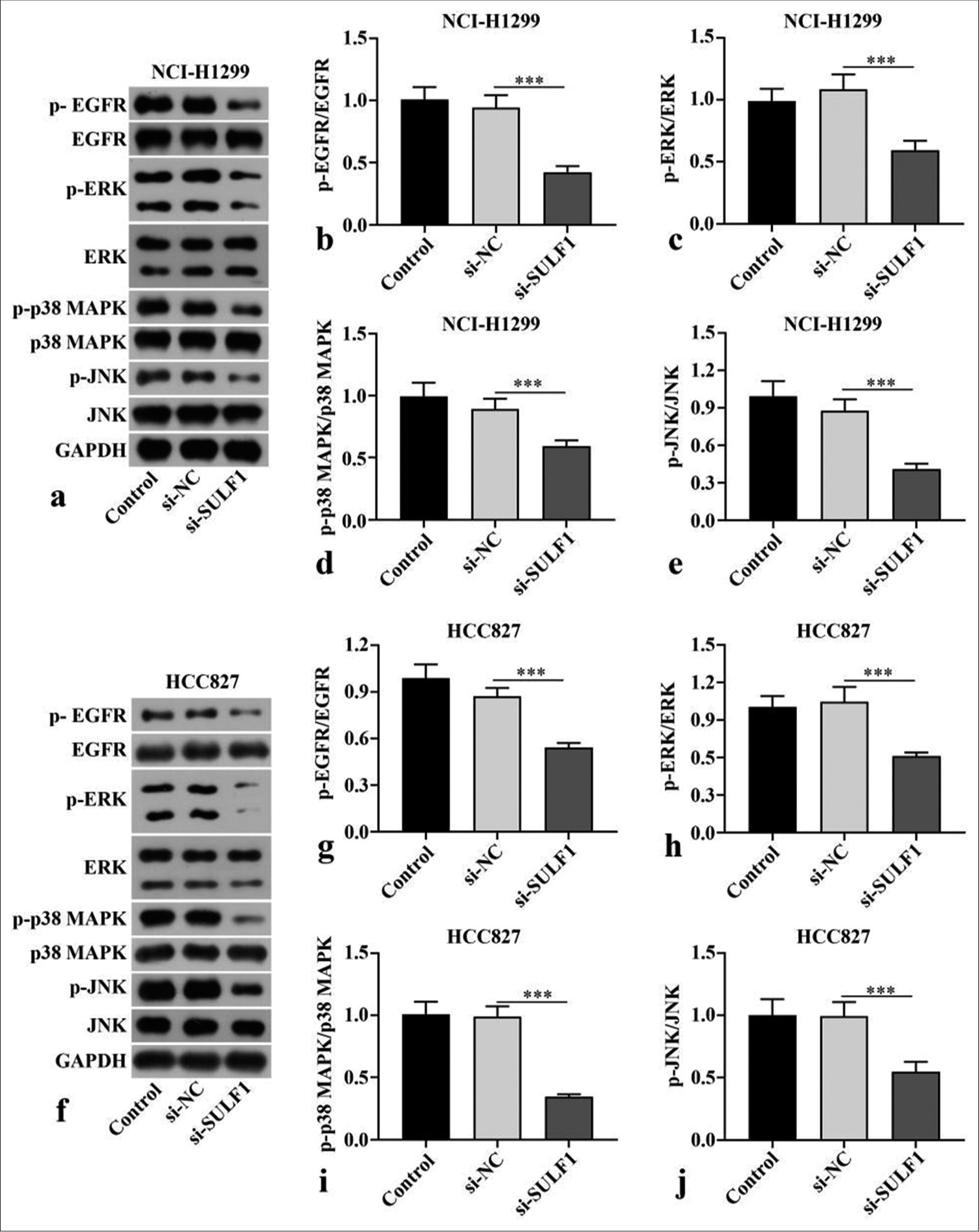
Export to PPT
EGFR/MAPK signaling pathway mediated the process of SULF1 knockdown suppressing metastasis in NCI-H1299 cellTo further confirm the mechanism of SULF1 in NSCLC cell metastasis, NCI-H1299 cells were handled with the EGFR/MAPK signaling pathway agonist NSC228155. Compared with the si-NC group, the si-SULF1 group showed prominently abated p-EGFR/EGFR, p-ERK/ERK, p-p38 MAPK/p38 MAPK, and p-JNK/JNK. Compared with the si-SULF1 group, the si-SULF1 + NSC228155 group showed considerably augmented p-EGFR/EGFR, p-ERK/ERK, p-p38 MAPK/p38 MAPK, and p-JNK/JNK protein levels in the NCI-H1299 cells (P < 0.01), [Figure 4a-c], as indicated by the Western blot results. The results from Transwell assay suggested that versus the si-NC group, the number of the migration and invasion cells of the NCI-H1299 cells in the si-SULF1 group was prominently reduced. Compared with the si-SULF1 group, the si-SULF1 + NSC228155 group showed considerably augmented cell migration and invasion (P < 0.05), [Figure 4d and e]. The E-cadherin protein level in the NCI-H1299 cells of the si-SULF1 group greatly increased relative to those in the si-NC group, whereas N-cadherin and vimentin protein levels were abated. The NCI-H1299 cells in the si-SULF1 + NSC228155 group showed abated E-cadherin protein level and considerably increased N-cadherin and vimentin protein levels relative to those in the si-SULF1 group (P < 0.05), [Figure 4f and g]. The protein levels were measured with Western blot.
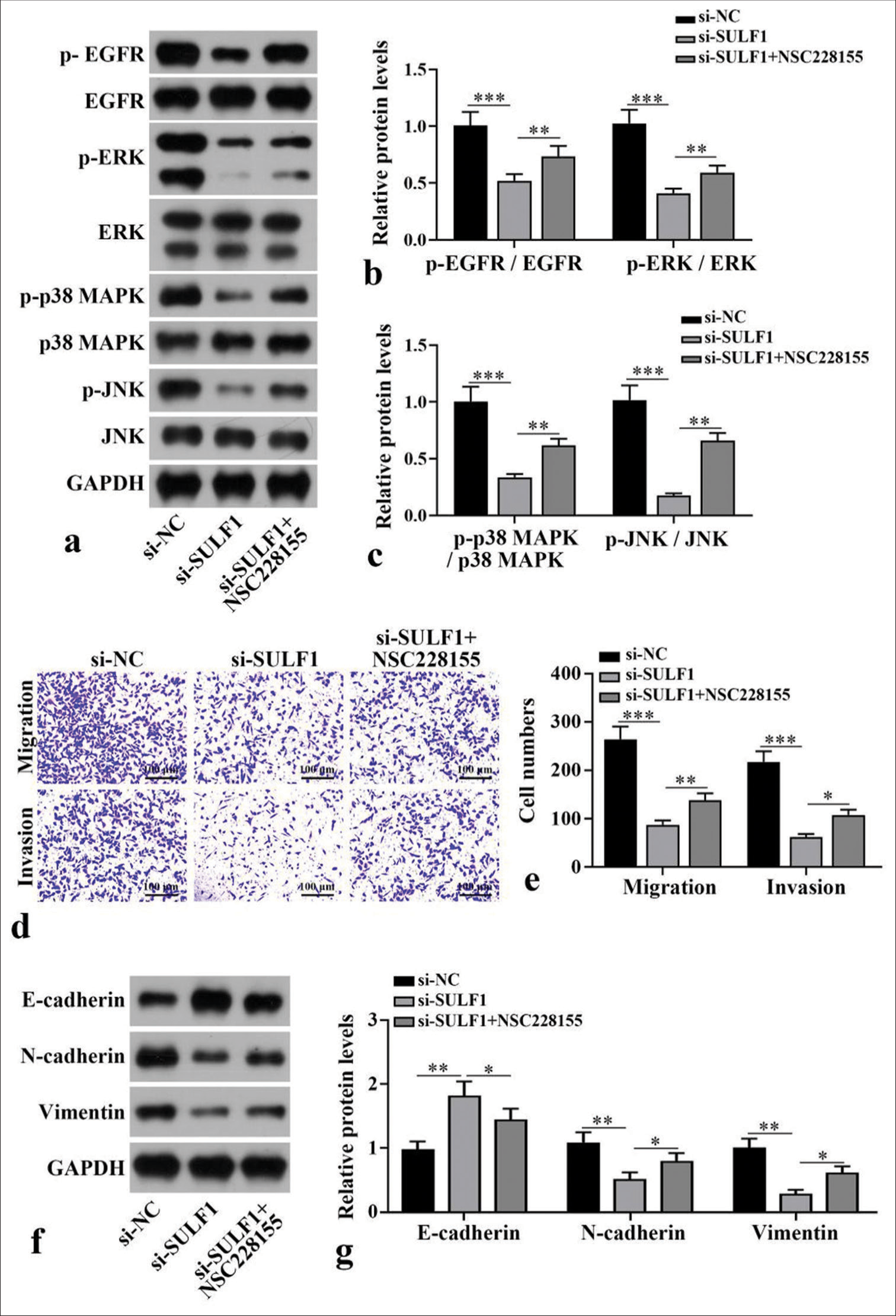
Export to PPT
High SULF1 expression was related to the poor prognosis of patients with NSCLCTo analyze the clinical characteristics of SULF1 and its relationship with the prognoses of NSCLC patients, we collected tumor tissues from 54 patients with NSCLC. A marked increase in SULF1 expression was observed in tumor tissues versus that in adjacent lung tissues (P < 0.001), [Figure 5a]. Table 1 displays the general information of 54 patients with NSCLC. No statistically significant difference between the high and low SULF1 expression groups was observed in terms of age, sex, pathological type, differentiation, and lymph node metastasis (P > 0.05), but TNM stage presented a significant difference between the two groups (P < 0.05), [Table 1]. The median survival times in the high and low SULF1 expression groups were 25 and 31 months, respectively. Furthermore, the overall survival in the high SULF1 expression group was lower than that in the low SULF1 expression group (P < 0.05), [Figure 5b], suggesting the poor prognoses of patients with NSCLC and high SULF1 expression levels.
Table 1: General information of patients with NSCLC.
NSCLC Patients (n=54) SULF1 expression χ2/t-value P-value High SULF1 expression (n=25) Low SULF1 expression (n=29) Age (year) 67.92±10.62 69.68±11.58 66.41±9.68 1.13 0.26 Sex (n, %) Male 34 (63.0) 16 (64.0) 18 (62.1) 0.02 0.88 Female 20 (37.0) 9 (36.0) 11 (37.9) Pathological type (n, %) Adenocarcinoma 30 (55.6) 14 (56.0) 16 (55.2) 0.34 0.84 Large-cell carcinoma 8 (14.8) 3 (12.0) 5 (17.2) Squamous cell carcinoma 16 (29.6) 8 (32.0) 8 (27.6) TNM stage (n, %) I–II 25 (46.3) 7 (28.0) 18 (62.1) 6.27 0.01 III 29 (53.7) 18 (72.0) 11 (37.9) Differentiation (n, %) Well and moderate 37 (68.5) 16 (64.0) 21 (72.4) 0.44 0.51 Poor 17 (31.5) 9 (36.0) 8 (27.6) Lymph node metastasis (n, %) Negative 31 (57.4) 14 (56.0) 17 (58.6) 0.04 0.85 Positive 23 (42.6) 11 (44.0) 12 (41.4)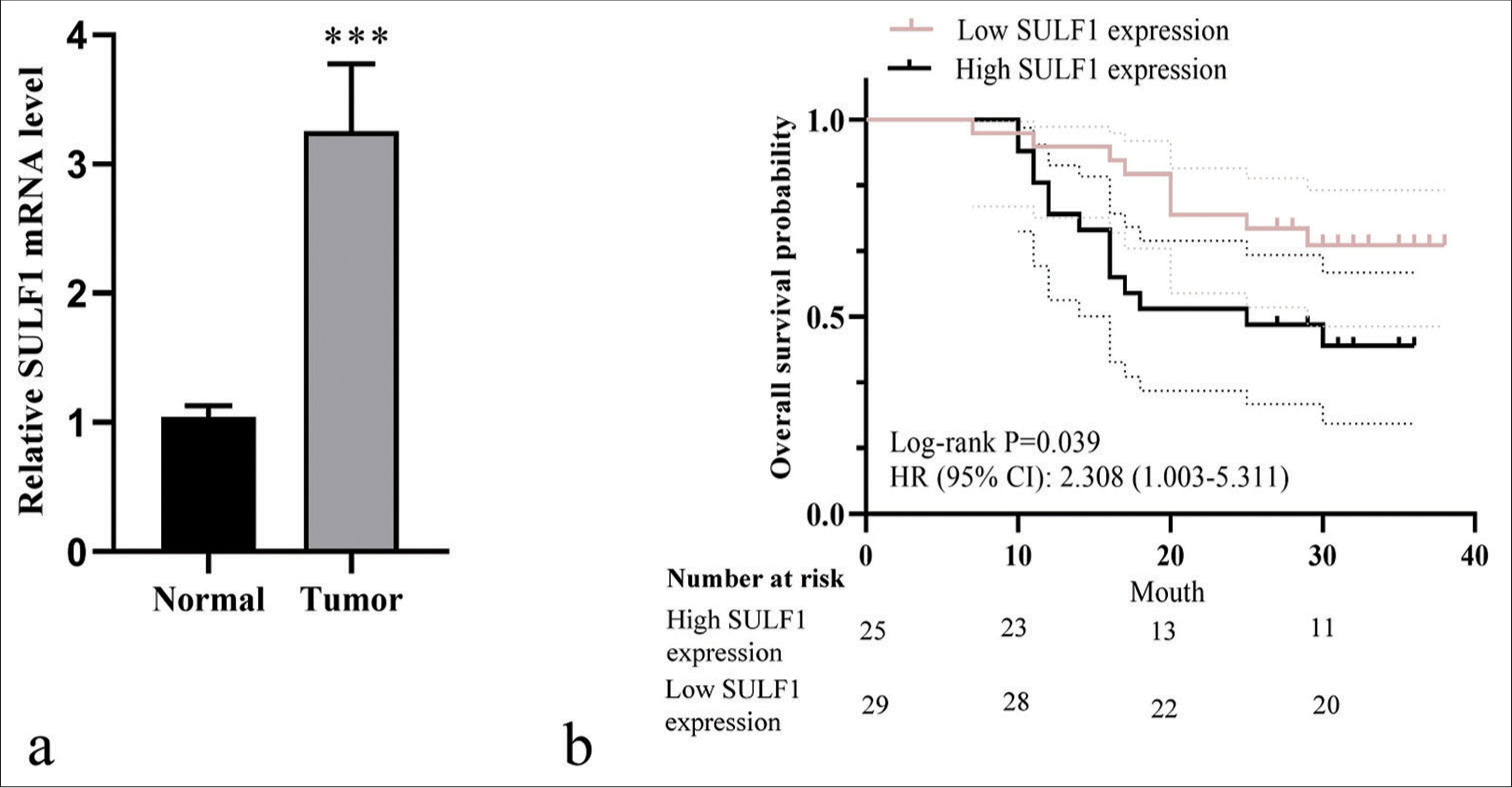
Export to PPT
DISCUSSIONSULF1 is highly expressed in several cancer types and regulates many signals by editing the sulfation modification of heparan sulfate proteoglycans.[15] However, understanding of SULF1 in NSCLC and its possible mechanism is lacking. Our study found that SULF1 level was strongly enriched in the NSCLC tissues and cells. Furthermore, patients with NSCLC and high SULF1 expression levels had poor overall survival. The knockdown of SULF1 abated the malignant biological behavior of the NSCLC cells, including proliferation, migration, and invasion.
EMT is an important cellular process that transforms polarized epithelial cells into mesenchymal phenotypes and increases cell motility.[16] EMT is abnormally activated under pathological conditions, including organ fibrosis and cancer. During EMT, the epithelial markers level is abated, whereas mesenchymal markers level is augmented.[17,18] The inhibition of EMT can help suppress the onset and advancement of NSCLC. Moreover, EMT contributes to chemotherapy and radiotherapy resistance.[19-21] The knockdown of SULF1 abated the biological behavior of the NSCLC cells, including migration, invasion, and EMT. Specifically, it upregulated the E-cadherin level and abated the N-cadherin and vimentin levels. These results indicated that knocking down SULF1 can inhibit the progression of NSCLC by regulating the EMT phenotype.
EGFR is related to a variety of complex cell signal transduction pathways. EGFR forms homodimers or heterodimers by binding to its ligands, thereby activating the PI3K/Akt and MAPK/ERK pathways in cancer.[22] In a study of colorectal cancer, the proliferation, migration, and invasion of colorectal cancer cells were promoted by the EGFR/MAPK pathway.[23] EGFR/MAPK signaling pathway is engaged in the regulation of malignant behavior in ovarian cancer cells, and blocking this pathway can repress the growth of xenograft tumors.[24] A study in lung cancer has shown that inhibiting the EGFR/MAPK pathway can restrain the growth and metastasis of lung cancer cells.[25] EGFR/MAPK was also linked to the osimertinib resistance in NSCLC.[26] In this study, the knockdown of SULF1 reduced p-EGFR/EGFR, p-p38 MAPK/p38 MAPK, p-ERK/ERK, and p-JNK/JNK levels. In addition, NSC228155 partially reversed the suppressive effect of SULF1 knockdown on NSCLC cells, indicating that the suppressive effect of SULF1 knockdown on NSCLC was exerted after the inactivation of the EGFR/MAPK pathway. For the limitations of this study, the relevant conclusions have not been confirmed through in vivo experiments, and the mechanisms of SULF1 and EGFR/MAPK signaling pathways have not been fully determined. Therefore, future studies will further explore the potential mechanism of SULF1 in the progression of NSCLC. In addition, NCI-H1299 and HCC827 were large-cell lung cancer and lung adenocarcinoma cell lines, respectively.
Squamous cell carcinoma cell lines, including NCI-H520 and H226, should be investigated as well.
SUMMARYThis study demonstrated the suppression of SULF1 knocking down on NSCLC. Mechanistically, SULF1 knockdown abated the invasion and metastasis of NSCLC through the EGFR/MAPK signaling pathway, suggesting that SULF1 is a therapeutic target for NSCLC.
AVAILABILITY OF DATA AND MATERIALSThe data used to support the current finding of this study are available from corresponding author upon request.
ABBREVIATIONSNSCLC – Non-small cell lung cancer
SULF1 – Sulfatase 1
EGFR – Epidermal growth factor receptor
MAPK – Mitogen activated protein kinase
EMT – Epithelial-to-mesenchymal transition
ERK – Extracellular signal-regulated kinase
PI3K – Phosphatidylinositol-3-kinase
LncRNA – Long non-coding ribonucleic acid
siRNA – Small interfering RNA
si-SULF1 – siRNA SULF1
si-NC – Negative control
qRT-PCR – Quantitative real-time polymerase chain reaction
GAPDH – Glyceraldehyde-3-phosphate dehydrogenase
CCK-8 – Cell Counting Kit-8
OD – Optical density
EdU – 5-Ethynyl-2-deoxyuridine
DAPI – 4',6-diamidino-2-phenylindole
JNK – c-Jun N-terminal kinase
p-JNK – Phospho-JNK
p-EGFR – Phospho-EGFR
p-ERK – Phospho-ERK
p-p38 MAPK – Phospho-p38 MAPK
TNM – Tumor node metastasis
PBS – Phosphate buffer saline
LUAD – Lung adenocarcinoma
HR – Hazard ratio
CI – Confidence interval
AUTHOR CONTRIBUTIONSTF and BLZ: Designed the research; BLZ, DPL, and LX: Performed the experiments;LX and JC: Analyzed the data. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Comments (0)