Oral squamous cell carcinoma (OSCC) is a prevalent malignant tumor.[1,2] Despite advancements in surgical techniques, radiotherapy, chemotherapy, biological therapy, and reconstructive procedures over the past three decades, the overall prognosis for oral cancer has insignificantly improved.[3] Cancer cells reprogram their metabolic pathways to sustain their energy and resource requirements. The Warburg effect highlights the preference of tumor cells for highly active aerobic glycolysis (AG) to meet their survival demands.[4,5] OSCC cells also exhibit distinct alterations in AG and glucose metabolism.[6,7] Understanding the mechanisms underlying AG in the development of OSCC offers novel avenues for therapeutic strategies.
Current research has identified Surfeit 4 (SURF4) as part of the Surfeit gene cluster.[8,9] SURF4 is a cargo receptor involved in transporting proteins between the endoplasmic reticulum and Golgi apparatus.[9] It also modulates the transport of apolipoprotein B (Apo B), which influences the transportation of very low-density lipoprotein (VLDL) precursors and regulates their secretion.[10] Liver-specific knockout of SURF4 has been shown to reduce cholesterol and triglyceride.[11] These findings indicate that SURF4 may affect protein and lipid metabolism, which presents potential clinical applications in cancer occurrence, development, and diagnosis. AG, which is a vital energy metabolism pathway, contributes to proliferation and invasion in cancer cells.
Our study aims to investigate the function and regulatory mechanism of SURF4 in oral cancer through cellular experiments and clinal samples. Specifically, we assessed the influence of SURF4 on proliferation, tumor stemness, invasion, and metastatic capacities in oral cancer cells. We, further, explored the regulatory role of SURF4 in AG and its effects on the metabolic pathways of cancer cells. These findings hold promise in providing innovative targeted strategies for oral cancer treatment. Gaining a deeper understanding of the functional and regulatory mechanisms of SURF4 can provide new insights and pave the way for oral cancer treatment and prevention.
MATERIAL AND METHODS Patient sample collectionThis study has been approved by the Medical Ethics Committee of Xingtai Medical College, (Approval No.: 2023029). The study began enrolling patients in September 2022 and ended in September 2023. Table 1 records the personal information and medical records of all participants. The inclusion criteria were as follows: patients who have not undergone chemotherapy or radiotherapy; patients who were diagnosed with OSCC; patients who are 18 years of age or older; and patients who provide informed consent to participate in the study. The exclusion criteria were as follows: patients who have received prior chemotherapy or radiotherapy; patients with a history of other malignancies; patients with severe systemic diseases or conditions that could interfere with the study; patients who are pregnant or breastfeeding; and patients who are unable or unwilling to comply with the study procedures. Participants were recruited for the negative control (NC) group, the OSCC group, and the OSCC with lymph node metastasis group. Sterile cotton swabs moistened with normal saline were used by dedicated hospital staff to collect oral mucosal epithelial cells or oral cancer cells from patients in a fasting state in the early morning. The freshly collected cell smears were centrifuged at a speed of 3000 rpm for 20 min at 4°C and stored in a −80°C freezer. Subsequently, Western blot analysis and quantitative reverse transcription polymerase chain reaction (qRT-PCR) were conducted to determine SURF4 levels in the cells. All patient samples were collected in compliance.
Table 1: Basic clinical characteristics.
No. Group Age Gender Pathological Stage Lymph node metastasis 1 Healthy 28 Male 2 Healthy 34 Female 3 Healthy 39 Female 4 Healthy 42 Male 5 Healthy 46 Male 6 Healthy 55 Male 7 OSCC 36 Male I stage No 8 OSCC 28 Female I stage No 9 OSCC 29 Male II stage No 10 OSCC 45 Female III stage No 11 OSCC 48 Female IV stage No 12 OSCC 53 Male III stage No 13 OSCC with LNM 31 Female IV stage Yes 14 OSCC with LNM 36 Male IV stage Yes 15 OSCC with LNM 39 Male IV stage Yes 16 OSCC with LNM 43 Female IV stage Yes 17 OSCC with LNM 48 Male IV stage Yes 18 OSCC with LNM 56 Female IV stage Yes Cells and reagentsThe OSCC cell strains squamous cell carcinoma (SCC)-25 (CRL-1628), CAL-27 (CRL-2095), Tac8113 (CRL-1623), and SCC-4 (CRL-1624) and human normal oral mucosal keratinocytes (hNOKs) were purchased from the ATCC cell bank (Manassas, Virginia, United States). Cells were cultured using Dulbecco’s modified Eagle medium (31600, Solarbio, Beijing, China). Before the experimentation, the cell lines were screened for mycoplasma contamination and validated for authenticity using short-tandem repeat analysis.
The selection criteria for the cell lines were based on their relevance to OSCC, which is the primary focus of this study. The cell strains SCC-25, CAL-27, Tac8113, and SCC-4 are well-established OSCC cell lines commonly used in research. These cell lines represent different subtypes of OSCC and exhibit characteristics similar to human OSCC tumors.
hNOKs were included as a control group to compare the behavior and response of OSCC cell lines with non-cancerous oral mucosal cells. This comparison helps identify specific changes and differences associated with the development and progression of OSCC.
The following reagents were purchased for the study: Restriction endonucleases (1022B), SYBR Premix Ex TaqTM II (639676), and reverse transcription reagents (RR037B) from Takara Company (Tokyo, Japan); plasmid extraction and gel recovery reagent kits (28706X4) from QIAGEN (Heidelberg, Germany); transfection reagent Lipofectamine 2,000 (11668500) and RNA extraction reagent Trizol (12183555) from Invitrogen (Carlsbad, California, USA); RIP (P0013B) and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel preparation kits (P0012A) from Beyotime Biotechnology (Shanghai, China); polyvinylidene fluoride (PVDF) membrane (IPFL00010) from Millipore (Bedford, Massachusetts, USA); and mouse anti-human b-actin (ab8226) from Abcam (Cambridge, MA, USA).
Cloning formation assayThe SCC-4 and Tac8113 cells (1,000 cells/well) were cultured in a 6-well plate for 24 h. They were then placed in a 5% CO2, 37°C incubator. Cloning formation size was observed from day 10 to day 14. The cells were gently washed with phosphate-buffered saline (PBS) for 3 times and air-dried. Next, 500 μL of 0.1% crystal violet was added to each well and incubated at 37°C for 30 min. Thereafter, the cells were washed with PBS. Finally, images were captured using a gel imaging system (ChemiDoc, Bio-Rad, Hercules, California, USA).
Cell adenosine 5’-triphosphate (ATP), extracellular acidification rate (ECAR), and lactate production assayCellular glucose consumption and lactate production were detected following the procedures outlined in reference.[12] The lactate (C0016) assay kits were purchased from Beyotime (Shanghai, China). Fluorometric Assay Kit for extracellular acidification rate ECAR (E-BC-F069) and chemiluminescence assay kit for ATP (E-BC-F002) were obtained from Elabscience (Wuhan, Hubei, China). Briefly, cells were enzymatically digested, counted, and plated in a 12-well plate overnight. After the culture medium was removed, fresh complete medium was added to each well, including blank control wells, followed by incubation for 48 h. The supernatant was collected, and the absolute cell number was counted. Lactate content was determined using a DADE BEHRING (RXL MAX) system (Deerfield, Illinois, USA). The levels of ATP and ECAR were analyzed through fluorescent enzyme labeling (Varioskan LUX, Thermo Fisher Scientific, Waltham, MA, USA).
Cell transfectionSCC-4 and TAC8113 were transfected using Lipofectamine 2000 transfection reagent with recombinant plasmids and empty vectors. First, 5 μL of lipid reagent and 4 μg of plasmid were added to 500 μL of culture medium and incubated separately. The two solutions were gently mixed. Cells were washed with the culture medium and added with the abovementioned mixture, followed by the addition of fresh culture medium.
Synthetic short hairpin RNA is a lab-made RNA molecule and was used for SURF4 regulation. Lentiviral vector GV493 was used for gene transfection. GeneChem (Shanghai, China) and Fenghui Biotechnology (Hunan, China) provided gene-related products and services. pLV-CMV-MCS-EF1-ZsGreen1-T2A-Puro was used as a lentiviral vector for gene expression and fluorescence labeling.
The sequence was synthesized by GeneChem Co., Ltd. The sense strand was 5’-AATTCAGCTTCCCTGGCTCT AGCAGCACAGAAATGGAAGCGAGTCGACATATTGC TGTTGCTGATGCTTCCATATTGGCTGTGCT GCTGTGCATGA-3’, and the antisense strand was 5’-CTAACTTGTCGACTCCCAGAACGGAA AACGTGTGCTGTGCGCTGGGTTTTGTGAG-3’. It was annealed to form a double-stranded DNA sequence and linked to the pcDNA3.1 vector using T4 DNA ligase to construct the SURF4 overexpression (OE) vector. The SURF4 knockdown sequences were as follows: the sense strand was 5’-CUCACAGAUCCUUUC-UAAGAUU-3’, and the antisense strand was 5’-UCUUAGAAGGAUCUGUGAGU-3’. Regarding the choice of specific concentrations for the OE and knockdown experiments, the concentrations were determined based on previous literature.[9] The aim was to achieve optimal expression or suppression of the target gene while minimizing any potential off-target effects or toxicity.
Thus, cells were divided into the control group, the NC (negative control) group, the SURF4 OE group, and the knockdown group, with six replicates for each group. The transfection efficiency was examined 48 h after transfection.
Cell counting kit-8 (CCK-8) assayCells were seeded in a 96-well plate, with three replicates for each group and approximately 5000 cells per well. At 0, 24, 48, and 72 h after seeding, 100 μL of culture medium (containing 10 μL of CCK-8 reagent [C0037, Solarbio, Beijing, China]) was added. Before measurement, bubbles were removed, and the optical density value at a wavelength of 450 nm (OD450) was detected using an microplate reader (ELx800, BioTek Instruments, Inc., Windim, Vermont, USA).
Transwell assay Cell migration assayTranswell inserts (S2391, BD Biosciences, San Jose, CA, USA) were inserted into a 24-well plate, and the bottom of each Transwell insert was coated with serum-free culture medium to form a chamber. The cells to be tested were suspended in serum-free culture medium (C11965500BT, Gibco, Grand Island, NY, USA) at an appropriate concentration. A certain amount of cell suspension was added to the upper chamber of the Transwell insert. The plate was placed in a cell culture incubator at 37°C for a period of time to allow the cells to migrate to the bottom of the insert. After incubation, the cells in the upper chamber were gently washed with PBS to remove non-migrated cells. The migrated cells at the bottom of the insert were fixed and stained for observation and counting. The number of migrated cells at the bottom of the insert was counted under a microscope (IX71, Olympus, Tokyo, Japan).
Cell invasion assayTranswell inserts were inserted into a 24-well plate, and the bottom of each Transwell insert was coated with a membrane containing matrix components (Beyotime, C0383, Shanghai, China). The cells to be tested were suspended in serum-free culture medium (C11965500BT, Gibco, Grand Island, NY, USA) at an appropriate concentration. A certain amount of cell suspension was added to the upper chamber of the Transwell insert (S2391, BD Biosciences, San Jose, CA, USA). The plate was placed in a cell culture incubator at 37°C for a period of time to allow the cells to invade into the bottom of the insert coated with invasion matrix. After incubation, the cells in the upper chamber were gently washed with PBS to remove non-invaded cells. The invaded cells at the bottom of the insert were fixed and stained for observation and counting. The number of invaded cells at the bottom of the insert was counted under a microscope (IX71, Olympus, Tokyo, Japan).
5-ethynyl-2’-deoxyuridine (EdU) incorporation assayThe 96-well plates were inoculated with 1 × 104 cells per well. Subsequently, 100 μL EdU (C0071S, Beyotime, Shanghai, China) was added to each well, followed by 50 μL of 4% paraformaldehyde after washing with PBS. The plates were incubated at room temperature for 30 min. After washing with PBS, 0.5% TritonX-100 (9002-93-1, Solarbio, Beijing, China) was added. The cell nuclei were then stained, and fluorescent imaging and image analysis were conducted using a high-content cell imaging system, Operetta (PerkinElmer, Waltham, Massachusetts, USA). The images were analyzed using ImageJ software (v1.8.0.345, National Institutes of Health, Bethesda, MD, USA).
Western blot analysisProteins were extracted using radioimmunoprecipitation assay lysate containing 1% phenylmethylsulfonyl fluoride (6542, Roche, Basel, Switzerland). Protein quantification was performed using the Bicinchoninic Acid Assay (BCA) kit (Cat #23225, Thermo Fisher Scientific, Waltham, MA, USA). A 20 μg protein sample was separated through 10% SDS-PAGE and then transferred to a PVDF membrane (IPVH00010, Millipore Corporation, MA, USA) using Western blot analysis. The membranes were blocked with 5% skim milk powder for 1 h at room temperature, followed by overnight incubation at 4°C with primary antibodies. The primary antibodies included polyclonal rabbit anti-SURF4 (1:1000, ab317407), anti-CD44 (1:1000, ab243894), anti-CD133 (1:1000, ab222782), anti-Hexokinase 2 (1:1000, ab209847), anti-Pyruvate kinase muscle 2 (1:1000, ab85555), anti- glucose transporter type 1 (1:1000, ab115730), anti- Lactate Dehydrogenase A (LDHA) (1:1000, ab52488), anti-phosphorylated adenosine monophosphate -activated protein kinase (AMPK-P) (1:1000, ab133448), anti-AMP-activated protein kinase (AMPK) (1:1000, ab32047), anti-glycogen synthase kinase 3 beta, (GSK-3b) (1:1000, ab93926), and anti-b-actin (1:8000, ab8226). The secondary antibody was horseradish peroxidase-conjugated to mouse anti-rabbit/mouse immunoglobulin G (1:5000, ab6728, ab6721). Primary and secondary antibodies were purchased from Abcam (Cambridge, MA, USA). The protein bands were observed by enhanced chemiluminescence (BL520b, Biosharp Life Science, Hefei, Anhui, China) and Image Quant LAS4000 (GE Healthcare, Chicago, IL, USA). Image J (v1.8.0, National Institutes of Health, Bethesda, MD, USA) was used to analyze strip gray values.
qRT-PCRTotal RNA was extracted using Trizol reagent (R0016, Beyotime Biotechnology, Shanghai, China), and reverse-transcribed into cDNA using the ABScript II complementary DNA (cDNA) first-chain synthesis kit (RK20400, ABClonal, Wuhan, Hunan, China). Reverse transcription of RNA into cDNA was conducted according to the instructions of the manufacturer. qRT-PCR was performed using Applied Biosystems’ 7500 Real-Time polymerase chain reaction (PCR) System (7500, Thermo Fisher Scientific, Waltham, MA, USA) and Synergy Brands Green PCR Master Mix (4312704, Thermo Fisher Scientific, Waltham, MA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control to determine the relative expression levels using the 2−ΔΔCt technique. The primer sequences (5’-3’) used in the study were follows: SURF4 F: ATGGGACAGAACGACCTGATG; R: GGTGTCGATATAGTCACGCTGOCT-4 F: AGAGG ATCACTTGGGGTACA; R:CGAAGCGACAGATGGT GGTCSOX-2 F:GCGGAGTGGAAACTTTTGTCC; R: GGG AAGCGTGTACTTATCCTTCT; GAPDH: F: GGT TGAGCA GGTACTTTT, R: AGCAAG AGCA CAAAGAGGAAG.
Statistical analysisStatistical analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, Inc., San Diego, California, USA), and one-way analysis of variance was conducted for comparison between groups. The difference between the two curves was compared and analyzed using a generalized equation. The results were reported as mean ± standard error of the mean. *P < 0.05 was considered statistically significant.
RESULTS Elevated serum SURF4 in OSCC: Clinical sample validationWe analyzed clinical samples from a hospital to validate the expression profile of SURF4 in different groups. A total of 18 patients were divided into three groups: the control group (n = 6), the OSCC group (n = 6), and the OSCC with lymph node metastasis group (n = 6). The three groups had no statistically significant difference in age. We observed a significant increase in SURF4 mRNA expression in OSCC patients compared with the control group (P < 0.001) [Figure 1a]. Western blot analysis also confirmed elevated levels of SURF4 in OSCC [Figure 1b and c]. Clinical samples confirmed that serum SURF4 levels were significantly higher in OSCC patients (P < 0.001). Moreover, we compared SURF4 expression in different states of OSCC and found a significant increase in the OSCC with lymph node metastasis group compared with the OSCC group (P < 0.01, P < 0.001). Western blot analysis and quantitative PCR yielded consistent results [Figure 1d-f]. Similarly, we examined SURF4 expression in common OSCC cell lines and found increased expression levels in SCC-25, CAL-27, Tac8113, and SCC-4 compared with human oral keratinocytes (HOKs) (P < 0.01, P < 0.001) [Figure 1g].
![Elevated serum SURF4 in OSCC: clinical sample validation. (a) Histogram showing the relative SURF4 mRNA expression levels in different clinical groups. (b) Western blot analysis of the expression of SURF4 proteins in each group. (c) Histogram showing the SURF4 expression levels in different clinical groups. (d) Histogram showing the SURF4 mRNA expression levels in each group (control, OSCC, and OSCC with lymph node metastasis). (e) Western blot analysis of the expression of SURF4 proteins in each group. (f) Histogram showing the SURF4 expression levels in each group (control group [n = 6], OSCC group [n = 6], and OSCC with lymph node metastasis group [n = 6]). (g) Histogram showing the SURF4 expression levels in different OSCC cell lines. (**P < 0.01; *** P < 0.001). (SURF4: Surfeit 4, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, OSCC: Oral squamous cell carcinoma, LNM: Lymph node metastasis, HOK: Human oral keratinocyte, OSCC cell lines: SCC-25, CAL-27, Tac8113, SCC-4: Squamous cell carcinoma-4, mRNA: Messenger RNA.)](https://cytojournal.com/content/105/2024/21/1/img/Cytojournal-21-60-g001.png)
Export to PPT
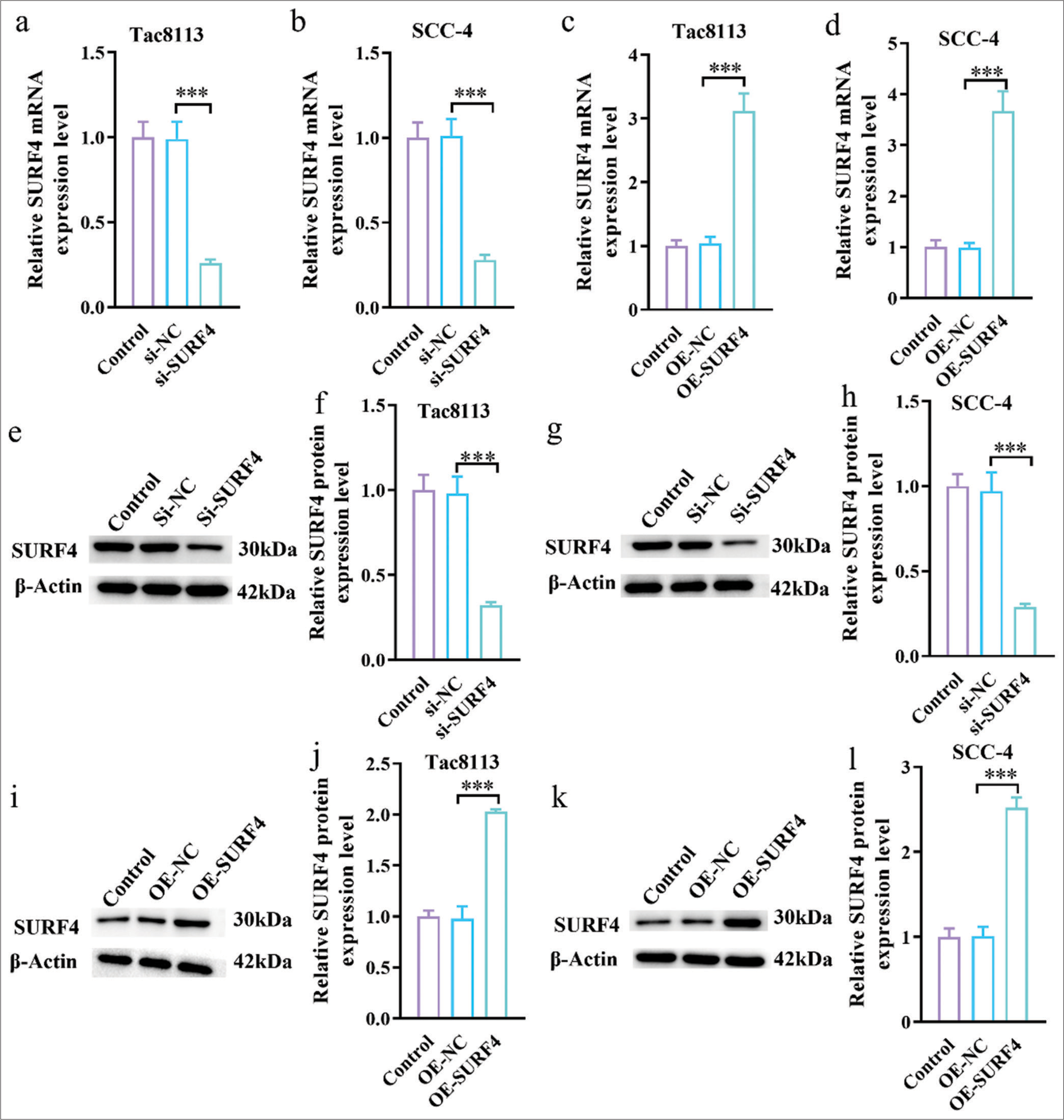
SURF4 regulates the proliferation and invasion abilities of OSCCsWe successfully transfected the SURF4 OE plasmid and small interfering RNA (SURF4) into the OSCC cell lines Tac8113 and SCC-4 to better understand the function of SURF4. As a result, stable cell lines with SURF4 OE and knockdown were established. qRT-PCR results showed that si-SURF4 significantly reduced the mRNA expression levels of SURF4 in Tac8113 and SCC-4 cells compared with the control and si-NC groups [Figure 2a and b] (P < 0.001). Conversely, OE-SURF4 significantly increased the mRNA expression levels of SURF4 in Tac8113 and SCC-4 cells compared with the control and OE-NC groups [Figure 2c and d] (P < 0.001). The efficiency of silencing and overexpressing SURF4 was further verified, as shown in Figure 2e-l. Western blot analysis results demonstrated successful silencing and OE of SURF4 in Tca8113 and SCC-4 cells.
Export to PPT
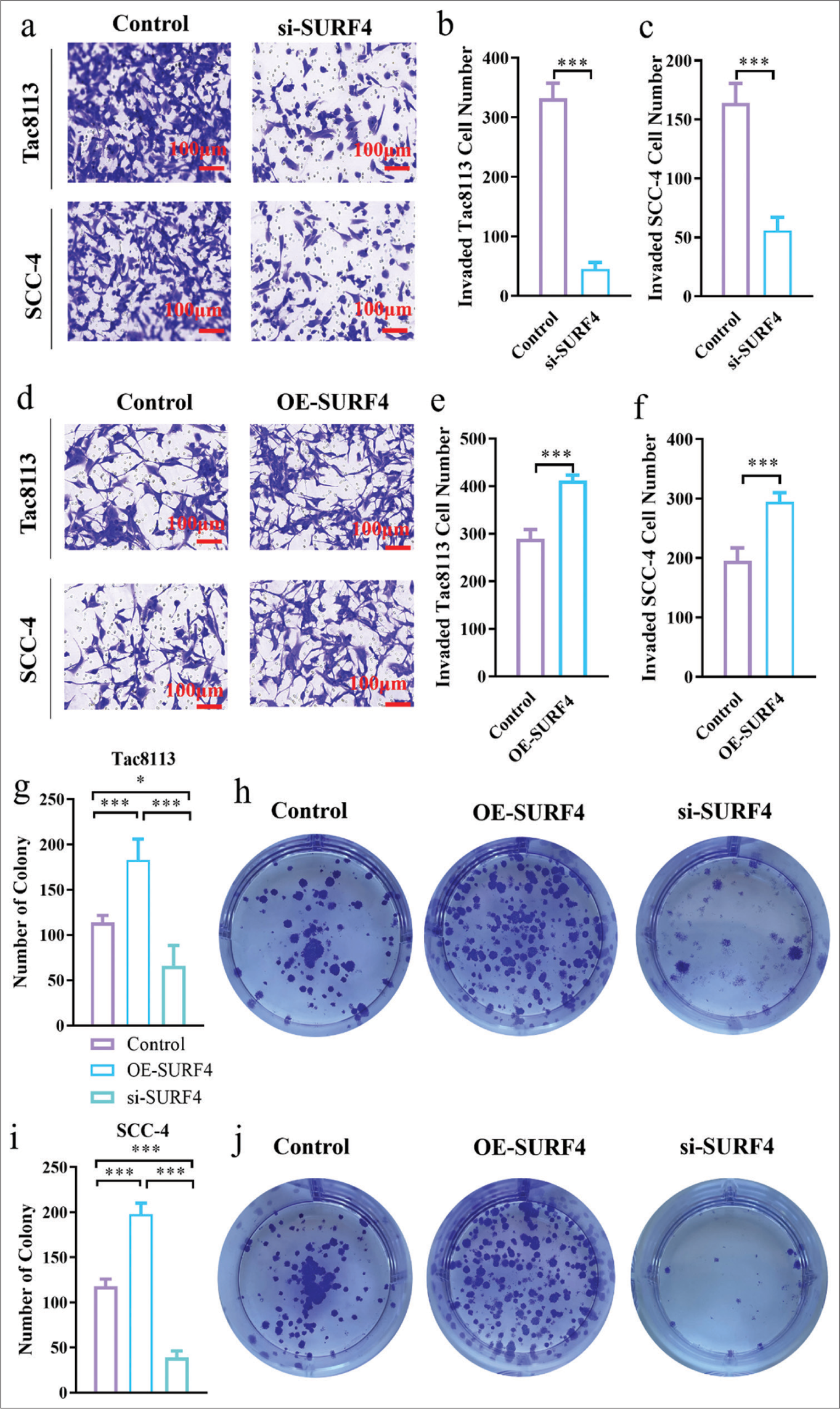
Transwell invasion assays were conducted to assess the effect of SURF4 on the invasion ability of OSCCs. We observed that SURF4 knockdown significantly reduced the invasion ability (P < 0.01, P < 0.001) in Tac8113 and SCC-4 cell lines [Figure 3a-c]. Conversely, the OE-SURF4 group exhibited increased invasion ability (P < 0.001) [Figures 3d-f]. Therefore, SURF4 plays a role in regulating invasion capability in oral cancer cells. Colony formation assays were performed to evaluate the proliferation ability of OSCCs. We observed a slight decrease in the total number of colonies after si-SURF4 (P < 0.05), while the OE-SURF4 group showed a significant increase (P < 0.001) [Figure 3g-j], which suggests enhanced proliferation ability in oral cancer cell lines with OE-SURF4. These functional experiments demonstrate the involvement of SURF4 in regulating the proliferation and invasion abilities of OSCCs.
Export to PPT
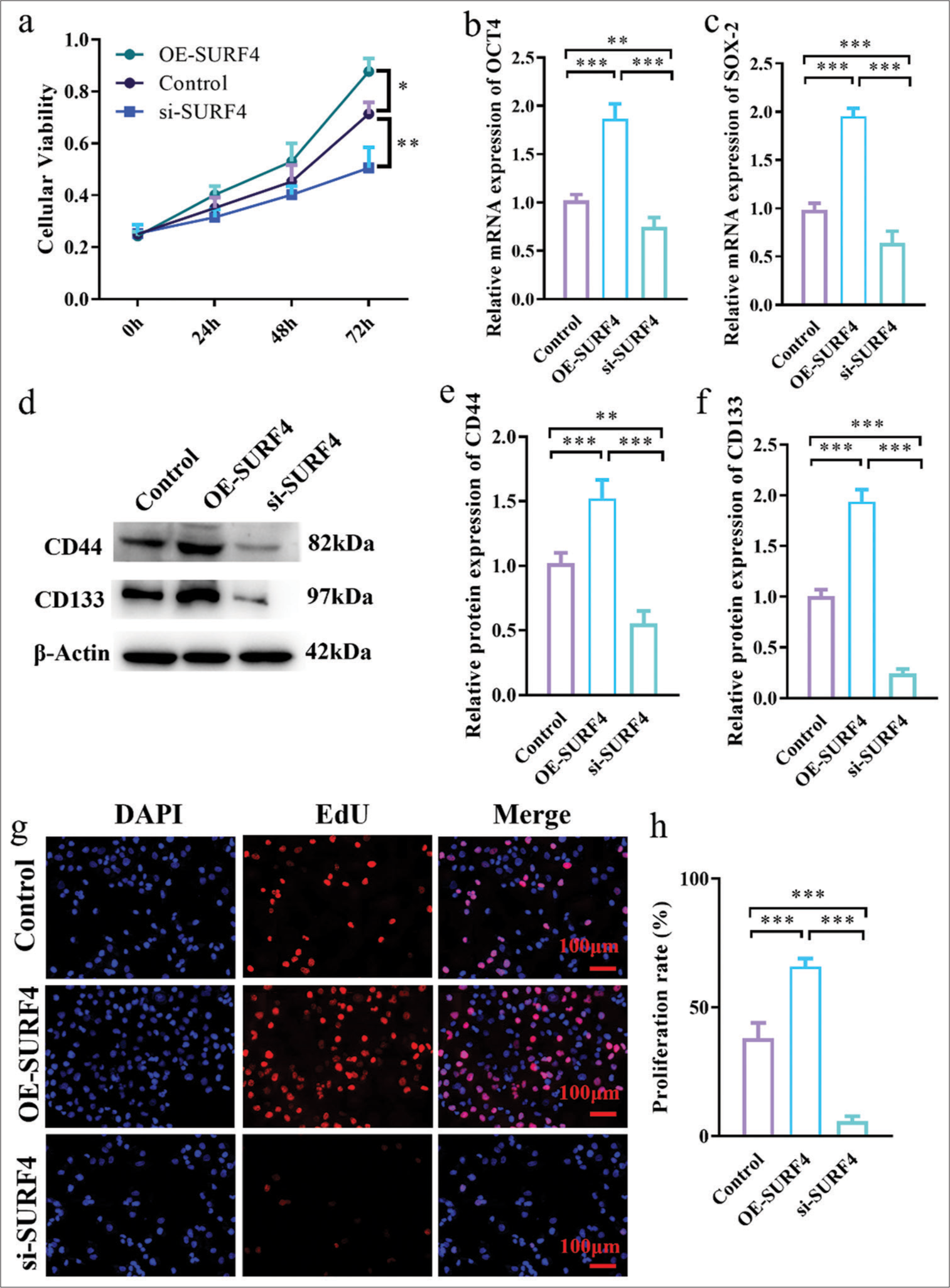
SURF4 aggravates the proliferation of OSCC and increases the level of stem cell genes OCT-4 and SOX-2CCK-8 assay results showed that, compared with the control group, the OE-SURF4 group significantly increased the viability of OSCC cells (P < 0.05), while the si-SURF4 group significantly decreased the viability of OSCC cells (P < 0.01) [Figure 4a]. Tumor stemness plays crucial role in the development and metastasis of oral cancer. To understand the role of SURF4 in tumor stemness, expression levels of stem cell genes OCT-4 and SOX-2 were compared in SCC-4, which reflected stemness. OE-SURF4 group showed higher expression of OCT-4 and SOX-2 (P < 0.001) [Figures 4b and c], whereas it exhibited lower expression after SURF4 knockdown (P < 0.01). Similarly, surface markers CD44 and CD133 exhibited consistent results. Increased expression of CD44 and CD133 was observed in the OE-SURF4 group (P < 0.001) [Figure 4d-f], while lower levels were found in the SCC-4 cell line after si-SURF4 (P < 0.05). These findings suggest that SURF4 may impact the proliferation and invasion of oral cancer through its effect on tumor stemness. Next, we employed EdU incorporation to validate the proliferative capacity of OSCCs (P < 0.001 control vs. OE-SURF4) [Figure 4g and h]. The results established that SURF4 may exert an influence on proliferation and invasion in oral cancer by impacting tumor stemness.
Export to PPT
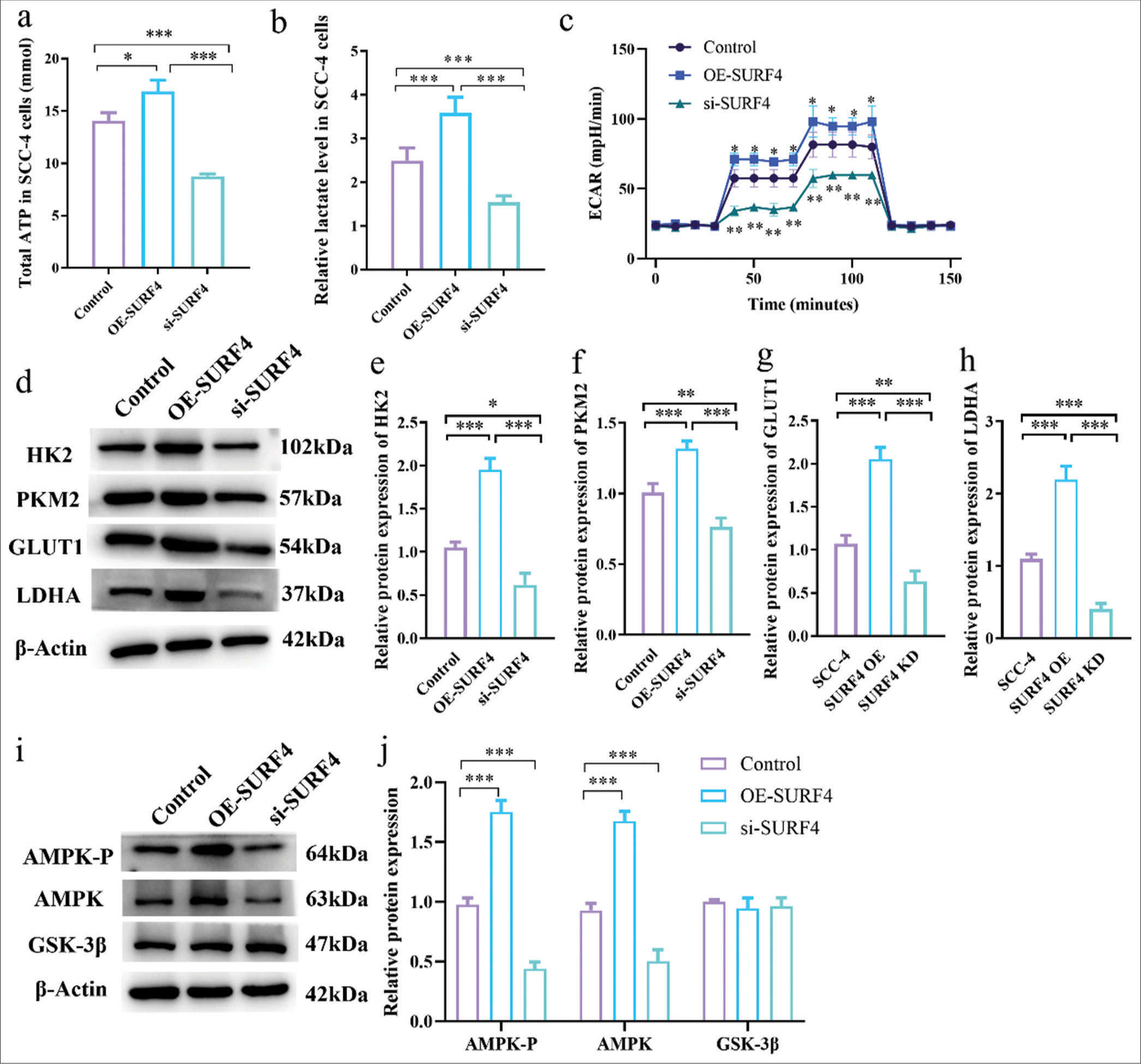
SURF4 impacts OSCC functionality by modulating AGResearch suggests that SURF4 is implicated in lipid transport and cellular metabolism, with tumor cells heavily relying on AG to sustain heightened proliferation. Therefore, we sought to ascertain the involvement of SURF4 in AG within oral cancer cells. Accordingly, we conducted AG assays on SCC-4 oral cancer cells in the OE-SURF4 and si-SURF4 groups. Remarkably, ATP levels (P < 0.05) [Figure 5a] and lactate levels [Figure 5b] (P < 0.001) were found to be elevated in the OE-SURF4 group, which implies an upregulated glycolytic state. Conversely, a notable reduction was observed in ATP and lactate levels (P < 0.001) on si-SURF4 in the SCC-4 cell line. Mitochondrial function, AG, and oxidative phosphorylation processes were evaluated through extracellular acidification rate (ECAR) measurements. The results revealed a significant increase in ECAR levels (P < 0.05) at various time points for the OE-SURF4 group, whereas ECAR levels displayed a considerable decrease (P < 0.01) in the si-SURF4 group [Figure 5c]. Furthermore, we assessed the expression of AG biomarkers, namely, hexokinase 2, pyruvate kinase M2, glucose transporter 1, and LDHA, as well as the AG-related regulatory proteins AMPK-P, AMPK, and GSK-3β. Western blot analysis unveiled elevated levels of AG-related regulatory proteins, except GSK-3b, in the OE-SURF4 group, while target proteins were diminished in the si-SURF4 group (P < 0.05, P < 0.01, and P < 0.001) [Figure 5d-j]. Overall, these findings substantiate the potential of SURF4 to impact the functionality of tumor cells by modulating AG.
Export to PPT
Inhibiting AG effectively impedes SURF4-induced promotion of proliferation and invasion in OSCCGiven that SURF4 can influence AG and subsequently facilitate tumor cell proliferation and metastasis, we postulated whether inhibiting AG could impede SURF4-mediated regulation of oral cancer cells. We introduced the AG inhibitor 2-deoxyglucose (2-DG) to investigate the role of AG in OSCC progression. The decision to use 2-DG as AG inhibitor was based on the previous study[13] supporting its efficacy in blocking glucose metabolism. 2-DG is a glucose analog that can enter cells and compete with glucose for phosphorylation by hexokinase, which inhibits AG. We introduced 2-DG to the OE-SURF4 group of SCC-4 oral cancer cells for validation, which resulted in a reduction in lactate levels (P < 0.001) [Figure 6a] and ECAR (P < 0.01) [Figure 6b]. Subsequent CCK-8 experiments were conducted to evaluate proliferation capacity in oral cancer cells, which revealed a diminished proliferative potential in the OE-SURF4 group following 2-DG treatment (P < 0.01) [Figure 6c]. Furthermore, when 2-DG was added, the protein levels associated with tumor stemness exhibited a decrease (P < 0.001) [Figure 6d]. Transwell (P < 0.01, P < 0.001) [Figure 6e and f] and EdU assays (P < 0.01, P < 0.001) [Figure 6g and h] were conducted to assess proliferation and invasion functionality in oral cancer cell. The analysis affirmed that inhibiting AG can effectively impede SURF4-induced promotion of proliferation and invasion in oral cancer cell.
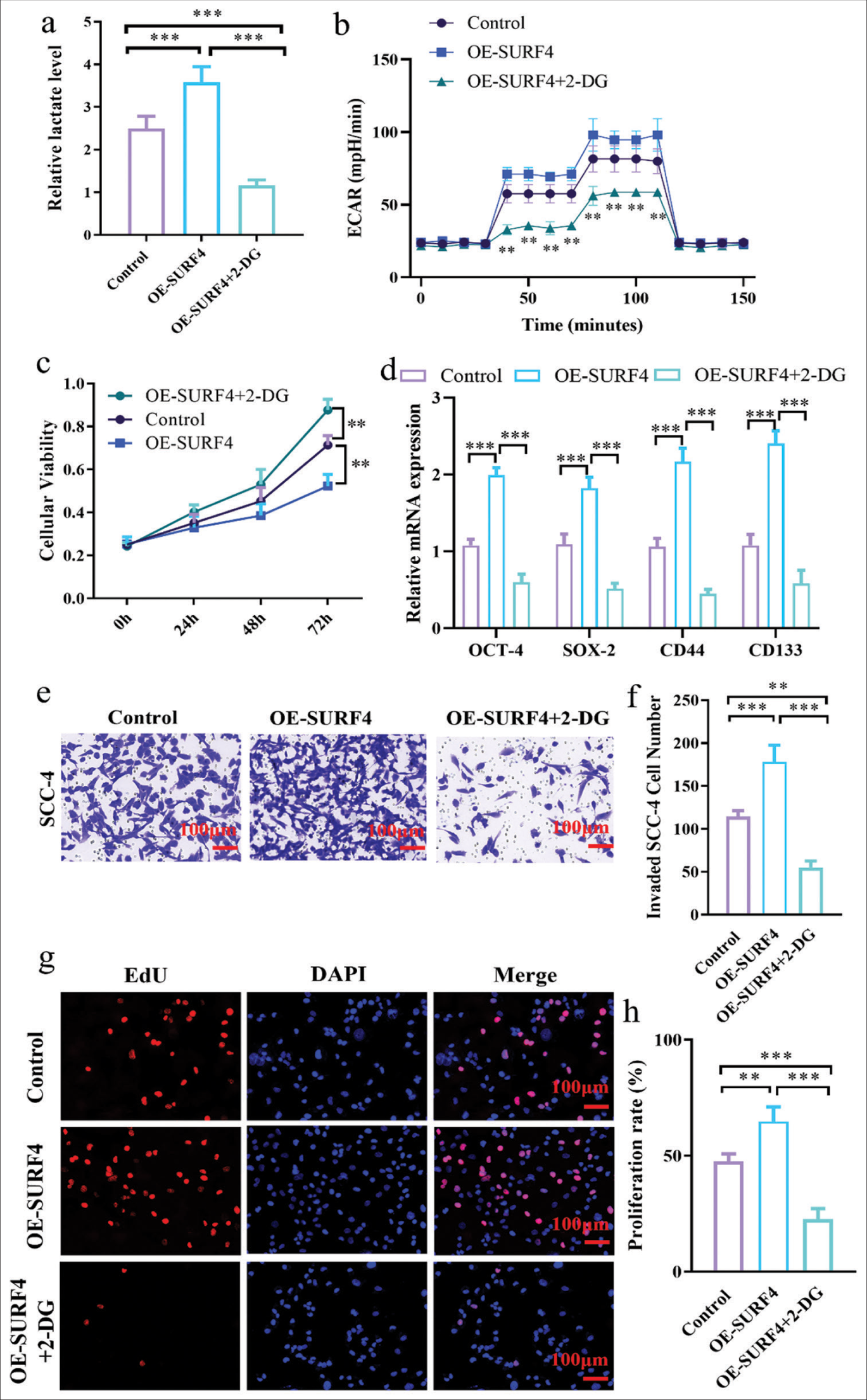
Export to PPT
DISCUSSIONOSCC is a prevalent malignant tumor, which accounts for 90% of head-and-neck malignancies.[14] The development of OSCC is a multifaceted process characterized by malignant cells exhibiting unlimited replicative capacity, apoptosis evasion, tissue invasion, and metastasis.[15] Cancer cells must reprogram their metabolic pathways to sustain the energy and molecular requirements necessary for acquiring and maintaining the aforementioned traits.[16] OSCCs also exhibit alterations in AG and glucose metabolism compared with normal cells.[17] These cancer cells preferentially rely on highly active AG, a phenomenon known as the Warburg effect, to meet their metabolic demands.
Recent research has elucidated the role of SURF4, which is a member of the Surfeit gene cluster.[18] SURF4 is widely expressed and localized to the endoplasmic reticulum membrane, which participates in the transport of various proteins between the endoplasmic reticulum and the Golgi apparatus. Furthermore, SURF4 is involved in the transport of Apo B, which impacts the transport of molecules of VLDL precursors and thus regulates their secretion.[19] Studies have shown that depletion of SURF4 significantly reduces plasma total cholesterol and triglyceride levels, which highlights its relevance in protein and lipid metabolism with potential clinical implications.[19,20] Another study has linked SURF4 to breast cancer,[21] which reinforces its association with cancer and suggests its potential as a therapeutic target in multiple cancer types. Discussing the mechanisms underlying AG in cancer progression can provide a more thorough understanding of the metabolic reprogramming observed in OSCC and its implications for cancer development and treatment.
Our findings underscore the pivotal role of SURF4 in the proliferation, tumorigenicity, invasion, and metastasis of oral cancer by modulating AG. OE of SURF4 in oral cancer cells leads to heightened ATP and lactate levels, augmented lactate production rates, and enhanced glycolytic activity. These conditions, in turn, foster cellular proliferation, invasion, and the maintenance of tumor stem cell populations. Conversely, inhibition of SURF4 suppresses lactate production and AG, which impedes the proliferation, tumorigenicity, invasion, and metastasis of oral cancer cells.
Some limitations and potential confounding factors can influence the outcomes of the study. One factor is the inherent variability in cell line models. While cell lines in our study are widely used as models for investigating OSCC, they may fail to fully recapitulate the complexity and heterogeneity of the actual patient population. Variations in genetic backgrounds, mutations, and cellular responses among different cell lines can potentially impact the results of the study. Another confounding factor to consider is the impact of different cell culture conditions. Cell culture techniques, such as the choice of growth media, supplements, and culture conditions, can influence cellular behavior and gene expression patterns. Variations in cultur
Comments (0)