Pancreatic cancer remains one of the most lethal malignancies and is characterized by an extremely low 5-year survival rate due to its aggressive nature and late diagnosis.[1,2] Common clinical treatment for pancreatic cancer is a combination of surgical resection and adjuvant chemotherapy.[3-5] Surgical treatment of pancreatic cancer has made significant advancements. Recently, three recent surgical approaches have improved the prognosis of patients with non-metastatic operable pancreatic cancer.[6] Advancements in modern imaging, radiotherapy techniques, and application of chemotherapy have significantly improved the control rates of pancreatic cancer. Despite these advances, the incidence of pancreatic cancer still increases. Treatment of pancreatic cancer can be further improved.[7]
Methyltransferase-like 14 (METTL14) usually acts as a N6-methyladenosine (m6A) methyltransferase in tumors and is closely related to tumor progression through methylation.[8-10] METTL14 may also affect the development of cancer by participating in the stability of RNAs.[11,12] Increasing studies have found that METTL14 could affect tumor progression through multiple mechanisms.[13] Wang et al.[14] illustrated that METTL14 could reduce PERP expression and promote tumor proliferation and cancer metastasis in pancreatic cancer. Chen et al. found that METTL14 deletion can remarkably reduce the lymphatic metastasis of pancreatic cancer.[15] Numerous canonical roles for the wingless-related integration site (Wnt)/β-catenin signaling system include tumor metabolism, embryonic development, cell fate determination, and the epithelial– mesenchymal transition. The Wnt pathway’s activation encourages b-catenin to build up in the cytoplasm and then go into the cell nucleus. In nucleus, b-catenin could act on T cell factor/lymphoid enhancer factor (TCF/LEF) promoters and regulate downstream genes of the Wnt pathway at the transcriptional level.[16] Moreover, c-Myc is commonly upregulated in pancreatic cancer and associated with its aggressive behavior.[17] As such, c-Myc has proved to be a target of METTL14.[18-20] This potential crosstalk among METTL14, c-Myc, and the Wnt/b-catenin pathway suggests a complex regulatory network that enhances pancreatic cancer cell proliferation and tumor progression.
Human pancreatic ductal cells and human pancreatic cancer cells were cultured in vitro to compare METTL14 level between pancreatic cancer and normal cells to verify our hypothesis and explore the function of METTL14 in pancreatic cancer. METTL14 expression in cells was regulated, and the cell proliferation rate, c-Myc expression level, and key protein activation status of the Wnt signaling pathway were compared. The c-Myc inhibitor 10058-F4 was used to determine whether METTL14 targets c-Myc in pancreatic cancer.
MATERIAL AND METHODS Pancreatic cell cultureNormal pancreatic ductal cell line (HPNE; Cat No. iCell-h102, iCell Bioscience Inc., Shanghai, China) and pancreatic cancer cell lines (PANC-1, Cat No. iCell-h172, iCell Bioscience Inc.; SW1990, Cat No. iCell-h203, iCell Bioscience Inc.) were purchased from iCELL Biotechnology Company (Shanghai, China). All the three cell lines were cultured in 1640 medium (Cat No. iCell-0002, iCell Bioscience Inc, Shanghai, China) with 5% carbon dioxide (CO2) at 37℃. The culture medium contained 10% fetal bovine serum (C0226, Beyotime, Shanghai, China) and 5% streptomycin/penicillin (15140122, Thermo Fisher Scientific, Waltham, MA, USA). Short tandem repeat testing was applied to authenticate the three cell lines, and mycoplasma testing was performed before using the cells to ensure that they are not contaminated.
Cell transfection and cell treatmentPlasmids encoding small interference RNA (si-RNA) against METTL14 (si-METTL14) or negative control (si-NC) were transfected into si-METTL14 group (si-METTL14 #1:5’-GGAUGAGUUAAUAGCUAAAUC-3’, 5’-UUUAGCUAUUAACUCAUCCUU-3’; si-METTL14 #2: 5’-GGAUGAUAUUAUGAAGUUAGA-3’, 5’-UAACU UCAUAAUAUCAUCCCA-3’; si-METTL14 #3: 5’-GACUGGUUGUACAGAAGAAAU-3’, 5’-UUC UUCUGUACAACCAGUCAA-3’) or si-NC group (5'-UUCUCCGAACGUGUCACGUTT-3' and 5'-A CGUGACACGUUCGGAGAATT-3') (GenePharma, Shanghai, China). The three si-RNAs were tested in our preliminary experiments, and the most effective one (si-METTL14 #1) was transfected in the si-METTL14 group. Plasmids encoding METTL14 (OE-METTL14, 5’-GTGCAGAAGGGACTAGGCAG-3’, 5’-GGGCACCC ACGTAATAGACC-3’) or blank plasmids (OE-NC) were transfected in the OE-METTL14 group or OE-NC group. All plasmids were designed by GeneChem (Shanghai, China), and Lipofectamine™ 3000 was used for transfection (L3000001, Invitrogen, CA, USA). The OE-METTL14+10058-F4 group was treated with c-Myc inhibitor 10058-F4 (Cat No. HY-12702, MedChemExpress, Monmouth Country, NJ, USA) after transfection of OE-METTL14.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) assayTRNzol Universal RNA simple Total RNA Kit (Cat No. DP424, Tiangen, Beijing, China) was used in total RNA extraction. Reverse transcription was conducted using FastQuant complementary DNA Synthesis Kit (Cat No. KR116, Tiangen). All measurements were analyzed using 2-ΔΔCT method. The sequence is shown in Table 1.
Table 1: The primer sequences.
Primer Primer sequences (5'-3') human METTL14 forward AGTTGGAACATGGATAGC human METTL14 reverse CTTCAACAATTATCGAGG mouse β-catenin forward TGACACCTCCCAAGTCCTTT mouse β-catenin reverse TTGCATACTGCCCGTCAAT mouse MMP-9 forward CCTGGAACTCACACGACATCTTC mouse MMP-9 reverse TGGAAACTCACACGCCAGAA mouse MMP-2 forward ACCCAGATGTGGCCAACTAC mouse MMP-2 reverse TACTTTTAAGGCCCGAGCAA mouse c-Myc forward GCGACTCTGAAGAAGAGCAAG mouse c-Myc reverse GCCTCGGGATGGAGATGAG GAPDH forward CAATGACCCCTTCATTGACC GAPDH reverse GATCTCGCTCCTGGAAGATG Western blot assayThe collected cells were mixed well with Radio Immunoprecipitation Assay (Cat No. P0013, Beyotime, Shanghai, China). The protein concentrations were measured using the bicinchoninic acid assay (BCA) kit (A045-4, Nanjing Institute of Bioengineering, Jiangsu, China) after the supernatant was collected. The soluble protein was denatured by heating it at 100°C for 5 min after being added at a ratio of one to ten to the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (BL529B, Biosharp Life Science, Hefei, Anhui, China). 15% SDS-PAGE (BL522A, Biosharp Life Science, Hefei, Anhui, China) electrophoresis was used to separate the denatured proteins. Targeted primary antibodies were incubated at 4°C while the separated proteins were transferred to the membranes (IPVH00010, Millipore Sigma, Billerica, MA, USA). All primary antibodies used in the research were listed as follows: Polyclonal METTL14 (1:1000 dilution; ab309096) and c-Myc (1:1000 dilution; ab185656) were obtained from Abcam (Cambridge, MA, USA). b-Catenin (1:2000 dilution, #8480), matrix metalloproteinase (MMP)-9 (1:2000 dilution, #3852), MMP-2 (1:2000 dilution, #40994), and Histone H3 (1:3000 dilution, #4499) were purchased from CST (Danvers, MA, USA) anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1000 dilution; PA1-987) was purched from Thermo Fisher Scientific (Waltham, MA, USA). Secondary antibody (1:2000 dilution; horseradish-peroxidase goat anti-rabbit immunoglobulin G; ab7090, Abcam, Cambridge, MA, USA) was incubated at room temperature. Protein bands were displayed with enhanced chemiluminescence (ECL) kits (BL520B, Biosharp Life Science, Hefei, Anhui, China) and Image Quant LAS4000 (GE Healthcare, Chicago, IL, USA) and analyzed by ImageJ software (version 2.0, LOCI, University of Wisconsin, Madison, Wisconsin, USA). Mean gray value of each protein band was recorded as raw measurement. The background signal for each band was subtracted to correct for non-specific staining. The intensity of each band was then normalized to a loading control (GAPDH or histone H3) to account for variations in protein loading or transfer efficiency.
Cell counting kit 8 (CCK8) assayCells were seeded in equal numbers into 96-well plates. Each detection hole contained 1.5 × 105 cells. Cell proliferation was assessed using the CCK8 (Cat No. G4103-1ML, Servicebio, Wuhan, Hubei, China) at 24-, 48-, 72-, and 96-h post-seeding. 15 μL of CCK-8 solution was applied to each well at the designated time and left for 2 h. Absorbance at 450 nm was recorded by a DR-200Bs reader (Diatek, Jiangsu, China).
Colony-forming assayCells were digested with pancreatic protein (Cat No. T1300, Solarbio, Beijing, China). A total of 1.5 × 103 cells were added to each well and cultured in an incubator at 37℃ and 5% CO2. Each well was stained with 0.1 % crystal violet (Cat No. C0121, Beyotime) dye. The number of clones was analyzed and counted using ImageJ software.
Wound scratch healing assayIn brief, 1.5 × 105 cells were seeded into a 12-well culture plate. Once the cells reached 100% confluency, a wound was created using a 20 μL pipette tip. Images were captured using an optical microscope at ×200 magnification (#ECLIPSE Ts2, Nikon, Tokyo, Japan) at the beginning of the assay and after 24 h of treatment. Migration ratio was then calculated using ImageJ software.
TdT-mediated dUTP-biotin nick end labeling (TUNEL) assayThe cells were fixed and permeabilized by 30 min of treatment with 4% paraformaldehyde and subsequent 5 min of treatment with 0.3% Triton X-100 (P0096, Beyotime, Shanghai, China), respectively. In the darkness at 37℃, the cells were incubated with terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) solution (C1089, Beyotime, Shanghai, China) for 60 min and 4’,6-Diamidino-2’-phenylindole (DAPI, BS097, Biosharp Life Science, Hefei, Anhui, China) for 15 min. DAPI is used to stain the nucleus. Four random fields were randomly selected for counting apoptotic cells using a fluorescence microscope (CKX53, OLYMPUS, Japan).
In vivo tumor growth assayTen six-week-old male Balb/c nude mice (18 ± 2 g) were obtained from the Jincheng General Hospital Animal Center (Jincheng, Shanxi, China). All animal experiments were conducted following the ethical guidelines approved by the Ethics Committee of Jincheng General Hospital (Approval No.:2024072301). PANC-1 cells (1 × 107) stably transfected with either the METTL14 knockdown vector or a control no-load lentivirus vector were injected into the right flanks of the mice. The mice were divided into shCtrl and shMETTL14 groups. After 5 weeks, the mice were euthanized by neck dislocation. The xenografted tumors were harvested for hematoxylin and eosin (HE) staining and immunohistochemical (IHC) analysis. Tumor volumes were calculated using the formula: volume = (length × width2)/2.
Lentivirus transfectionLentiviral vectors containing shCtrl (pLKO.1) and shMETTL14 (5'-ACCTCGCCGTGTTAAATAGCAAAGAT TCAAGAGATCTTTGCTATTTAACACGGCTT-3') were constructed by GenePharma (Shanghai, China) and utilized for cell transfection. PANC-1 cells were transfected for 48 h. The cells were cultured in medium containing 5 μg/mL puromycin (P8833, Sigma–Aldrich, Billerica, MA, USA) for 2 weeks to select for stably transfected cells. The verified pools of knockdown cells were then utilized for subsequent experiments.
HE staining and IHC analysisThe tumor tissues were sectioned to a thickness of 4 μm after being fixed in paraffin. The tissue samples were then stained with HE or incubated with the specified primary antibodies using the Elivision™ plus polymer horseradish peroxidase immunohistochemistry kit (D41-18, Maxim, Fujian, China).
For HE staining, hematoxylin-eosin stain kit (G1120, Solarbio, Beijing, China) was used.
For IHC assay, the tumor sections incubated with the primary antibody Ki-67 (1:1000 dilution, #9449S, CST, Danvers, MA, USA).
The staining images were observed by microscope (CKX53, OLYMPUS, Tokyo, Japan), and lung metastasis was quantified. Lung metastasis were evaluated by HE staining. Ki-67 was statistically analyzed by ImageJ software (v1.8.0.345, National Institutes of Health, Bethesda, MD, USA).
Immunofluorescence stainingImmunofluorescence was used to locate b-catenin. After being fixed in 4% paraformaldehyde, the cells were injected into phosphate-buffered saline for 10 min with 0.1% Triton X-100. The cells were sealed with 2% bovine serum albumin for 15 min. Primary antibody b-catenin (1:200 dilution, #8480T, CST, Danvers, MA, USA) was incubated at 4°C overnight. The nuclear were stained with DAPI (C1002, Beyotime Biotechnology, Shanghai, China). Finally, laser scanning confocal microscopy (LSM900, Zeiss, Oberkochen, Baden-Württemberg, Germany) was used for imaging.
Statistical analysisThe Statistical Package for the Social Sciences 20.0 software was used for data analysis (Analysis Software Co., Ltd., Shanghai, China). All the data were presented as mean ± standard deviation and assessed by independent two-tailed Student’s t-test. Multiple comparisons between multiple sets of data were conducted using one-way analysis of variance with Tukey’s test for post hoc analysis. Data analysis was performed using GraphPad 8.0.2 software (GraphPad Software, La Jolla, CA, USA). Statistical significance was considered at P < 0.05.
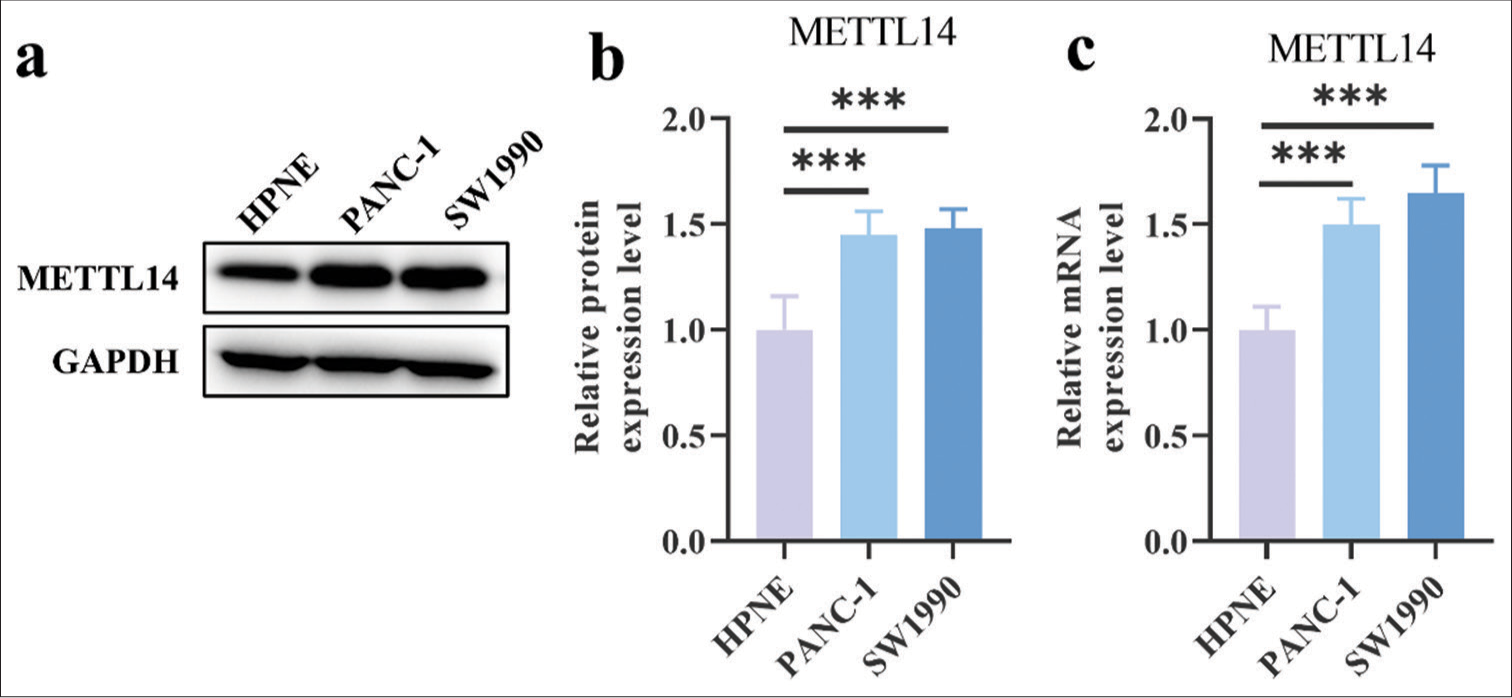
RESULTS Expression of METTL14 in different cellsThe protein levels of METTL14 in PANC-1 (P < 0.001) and SW1990 (P < 0.001) were higher than that in HPNE [Figure 1a and b]. Furthermore, METTL14 messenger (mRNA) had higher expression in PANC-1 [Figure 1c], (P < 0.001) and SW1990 [Figure 1c], (P < 0.001) than in HPNE.
Export to PPT
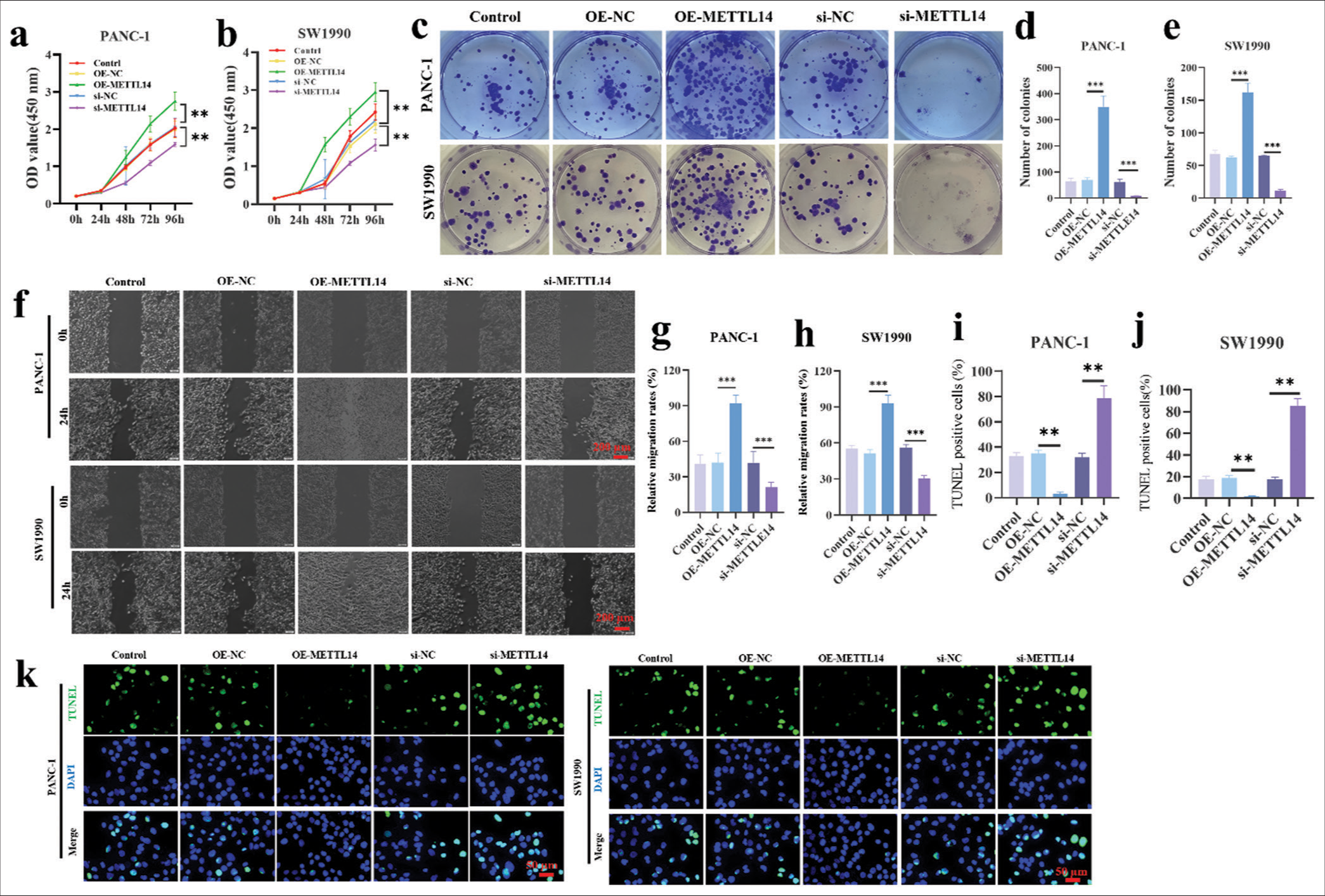
METTL14 promoted the proliferation and migration of pancreatic cancer cellsThe CCK8 assay results in transfected PANC-1 cells [Figure 2a] and SW1990 cells [Figure 2b] demonstrated that the cell viability increased in OE-METTL14-transfected cells (P < 0.01) but decreased in si-METTL14-transfected cells (P < 0.01). Figure 2c-e showed that the colony formation number of OE-METTL14-transfected cells (P < 0.001) increased; this number in si-METTL14-transfected cells (P < 0.001) decreased. In the results of wound scratch healing assay [Figure 2f-h], the cancer cell migration ability increased in OE-METTL14-transfected cells (P < 0.001) but decreased in si-METTL14-transfected cells (P < 0.001). In Figure 2i-k, OE-METTL14-transfected groups (P < 0.01) had less apoptotic pancreatic cancer cells than OE-NC groups. The si-NC groups had less apoptotic pancreatic cancer cells than the si-METTL14-transfected group (P < 0.01). These results showed that METTL14 promoted the tumor proliferation and migration of pancreatic cancer.
Export to PPT
METTL14 promoted c-Myc expression and nucleus β-catenin expression in pancreatic cancer cell linesThe protein expression of key factors such as c-Myc, b-catenin, MMP-9, and MMP-2 was explored. Figure 3a-g showed that the expression of c-Myc, MMP-2, and MMP-9 increased iafter METTL14 treatment and decreased after silencing METTL14. Interestingly, total expression of b-catenin remained unchanged in the OE-METTL14 group and si-METTL14 group, while nucleus b-catenin protein expression was increased after METTL14 overexpression and decreased aftern silencing METTL14 (P < 0.01). The results of b-catenin immunofluorescence staining are shown in Figure 3h and i. The positive b-catenin signal was strong in the OE-METTL14 group but weak in the si-METTLE14 group. All these results indicated that METTL4 could promote the protein expression of c-Myc, MMP-9, and MMP-2, but not b-catenin. METTL4 may induce the movement of b-catenin into the nucleus and activate b-catenin signaling.

Export to PPT
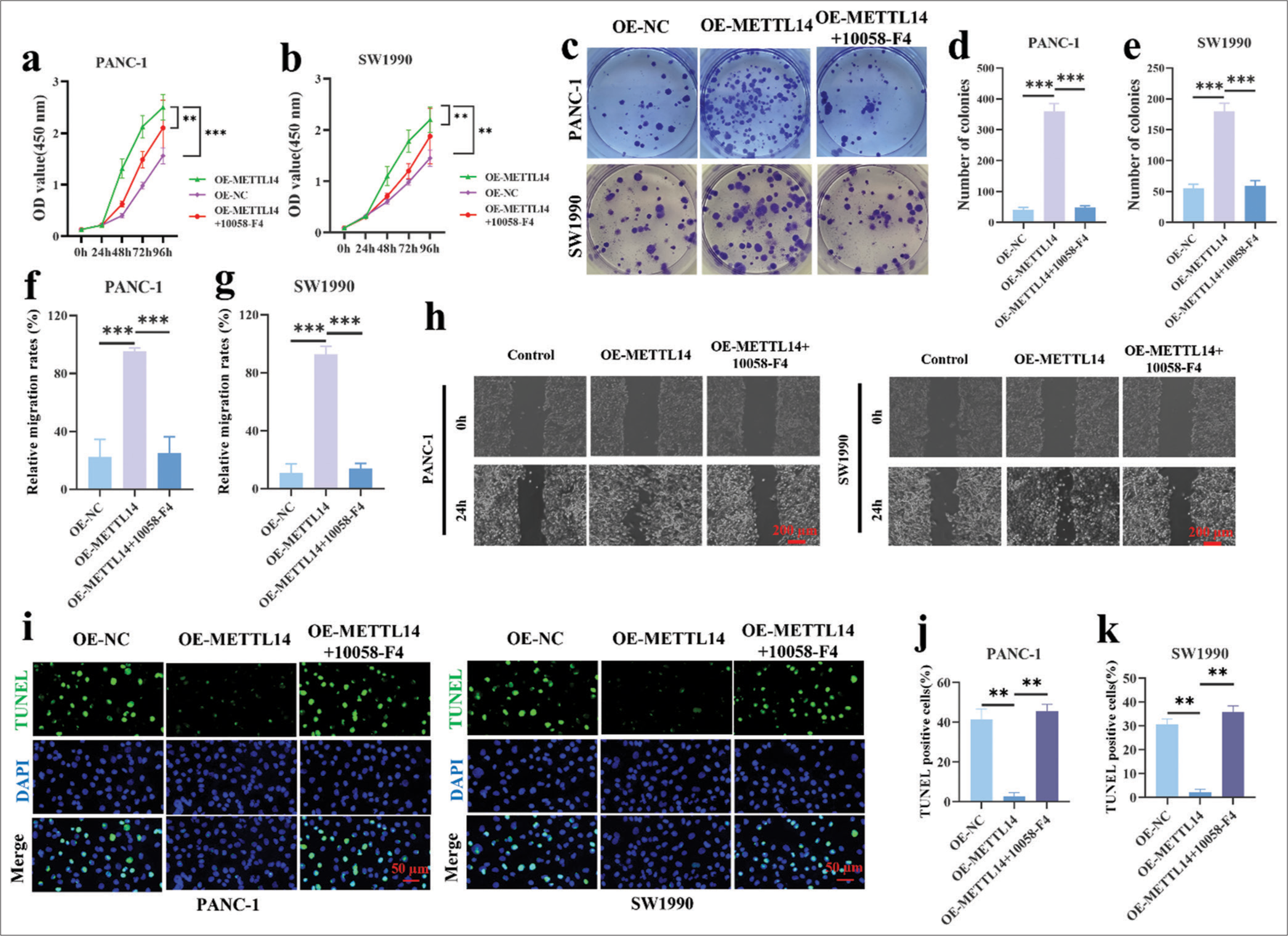
Effect of c-Myc inhibitor (10058-F4) on METTL14 overexpressed pancreatic cancer cellsThe CCK8 assay [Figure 4a] showed that PANC-1 cells in the OE-METTL14 group proliferated more than cells in the OE-NC group at 96 h (P < 0.001), and this effect was reversely inhibited by 10058-F4 (P < 0.01). Figure 4b demonstrates that the cell proliferation of SW1990 increased at 96 h by OE-METTL14 transfection (P < 0.01) and reversely decreased by 10058-F4 (P < 0.01). As shown in Figure 4c-e, the increased colony formation ability of OEMETTL14-transfected cells (P < 0.001) was reversed by 10058-F4 (P < 0.001). In the results of wound scratch healing assay [Figure 4f-h], the increase in cell migration ability in OE-METTL14-transfected cells (P < 0.001) was reversed by 10058-F4 (P < 0.001). In Figure 4i-k, 10058-F4 treatment also increased the apoptotic cell rate in the OE-METTL14-transfected group (P < 0.01). All these results proved that OE-METTL14 upregulated METTL14 expression and promoted the proliferation of pancreatic cancer cell lines PANC-1 and SW1990 by upregulating c-Myc. Otherwise, the effect of upregulating METTL14 on promoting pancreatic cancer cell proliferation could not be fulfilled.
Export to PPT
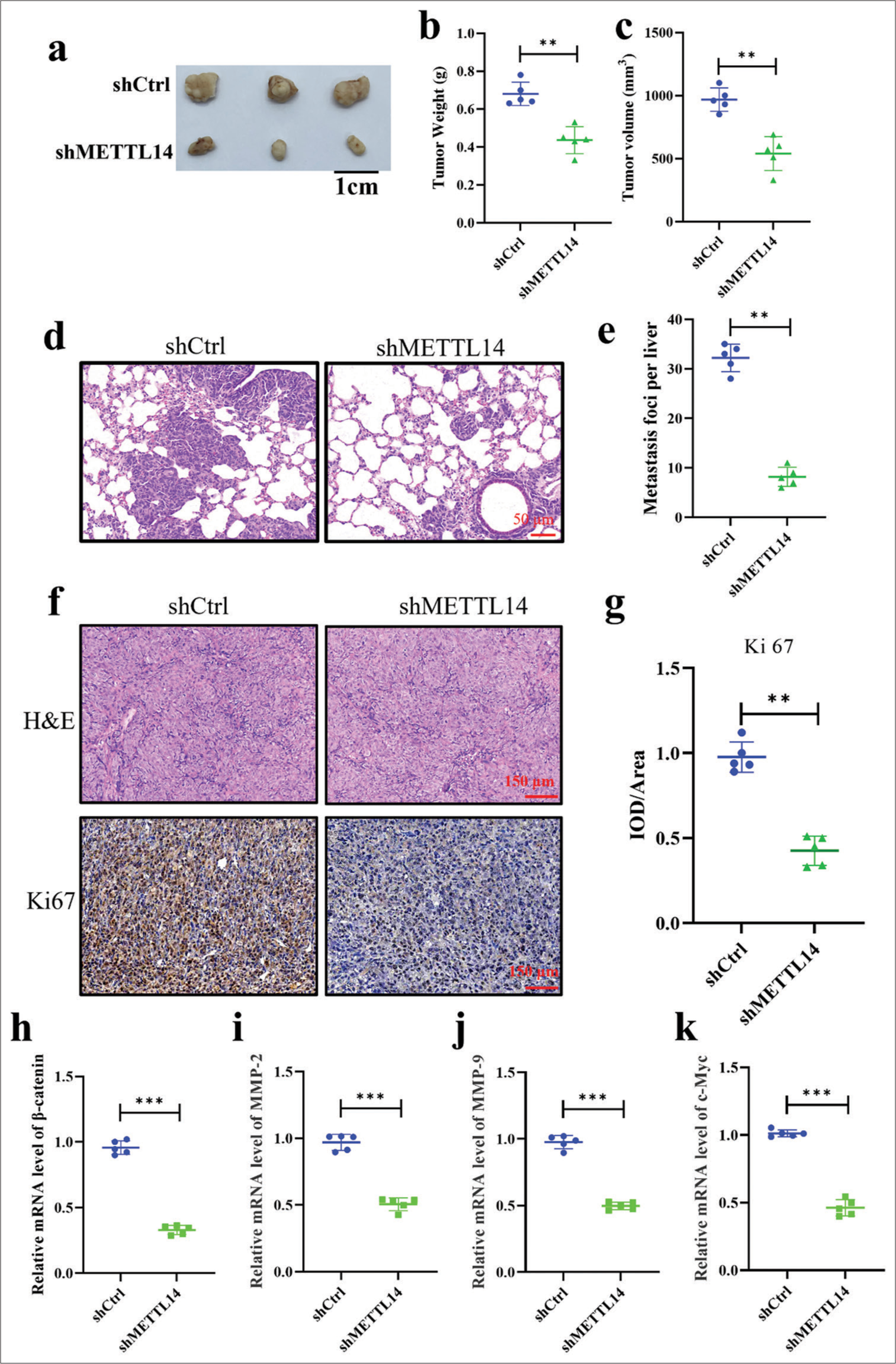
METTL14 promoted tumor growth and metastasis of PANC-1 in vivoTo investigate the potential of METTL14 in promoting tumor growth in vivo, we generated a stable METTL14 knock down PANC-1 cell line (shMETTL14) and a corresponding negative control (shCtrl) cell line. After 35 days of observation, xenograft tumor volume and weight decreased significantly after silencing METTL14 [Figure 5a-c] (P < 0.01). The METTL14 knockdown group had significantly inhibited PANC-1 cell metastases to the lung [Figure 5d and e], (P < 0.01). Figure 5f and g revealed a downregulation of Ki-67 in the METTL14 knockdown tumors compared with the controls (P < 0.01). In addition, shCtrl cells were loosely arranged with gaps. After silencing METTLE14, the cells were tightly arranged. Finally, the mRNA expression of b-catenin, MMP-2, MMP-9, and c-Myc was determined by qRT-PCR [Figure 5h-k]. The results of animal experiments were similar to those of PANC-1 cells. The mRNA expression levels of MMP-2, MMP-9, b-catenin, and c-Myc significantly decreased after silencing METTLE14 (P < 0.001).
Export to PPT
DISCUSSIONThe proliferation of pancreatic cancer cells is a critical aspect in tumor growth and malignancy. When pancreatic cancer occurs, the cancer cells will proliferate and their invasion and anti-apoptotic ability will increase.[21,22] Our results demonstrate that METTL14 is preferentially upregulated in pancreatic cancer cell lines. The overexpression of METTL14 in these cancer cell lines significantly enhanced their proliferation and colony-forming capabilities, as evidenced by the CCK8 and colony formation assays. Silencing METTL14 led to a marked reduction in these proliferative activities. The TUNEL assay results also indicated that METTL14 overexpression decreased in pancreatic cancer cells. These observations are consistent with a previous report that METTL14 acts as a promoter of tumorigenesis in pancreatic cancer.[14] In addition, the downregulation of METTL14 increased apoptosis and autophagy induced by cisplatin in pancreatic cancer cells.[23] We further explored the mechanism of action of METTL14 in pancreatic cancer.
The progression of pancreatic cancer is a dynamic process that involves multiple gene expression, modification, and cytokine activation.[24-27] c-Myc and beta-catenin are particularly linked to pancreatic cancer. Previous reports found that pancreatic cancer metastasis is promoted by activating c-Myc transcription.[28] b-catenin may be the target of miRNAs involved in pancreatic cancer development.[29] A critical observation from this study is the effect of METTL14 on c-Myc signaling and b-catenin. The total expression of b-catenin did not change, but the overexpression of METTL14 led to increased b-catenin expression in the cell nucleus. This translocation of b-catenin into the nucleus is crucial because it facilitates the transcription of c-Myc, MMP-9, and MMP-2, which are associated with cell proliferation and metastasis.[16,17,30] The upregulation of these key factors underscores the role of METTL14 in promoting a pro-tumorigenic environment through c-Myc signaling.
Furthermore, this study employed the c-Myc inhibitor 10058-F4 to determine the relationship between METTL14 and c-Myc. Many studies have shown that 10058-F4 is a reliable c-Myc inhibitor.[31-33] The inhibition of c-Myc effectively reversed the proliferative advantages conferred by METTL14 overexpression, suggesting that c-Myc is a pivotal target of METTL14 in the context of pancreatic cancer cell proliferation. This finding indicates c-Myc as a central regulator of cell growth and a common oncogene in pancreatic cancer. We further discussed the role of c-Myc in lung adenocarcinoma by combining METTL14 with c-Myc inhibitors. However, this study has some limitations. This study only used animals and cells for experiments, and the types of experimental samples were not rich enough. In the follow-up in-depth investigation, clinical samples of patients may be used to further prove our conclusions.
This study found that METTLE14 alleviates the occurrence and metastasis of lung adenocarcinoma by affecting the expression of c-Myc and activating b-catenin into the nucleus.
SUMMARYThis research highlights the significant role of METTL14 in the proliferation and development of pancreatic cancer. By enhancing c-Myc expression, METTL14 facilitates a cellular environment conducive to tumor growth and survival. These results not only advance our knowledge of the molecular pathways driving the advancement of prostate cancer but they also point to METTL14 as a possible therapeutic target for the creation of novel therapeutic approaches. Further investigations are warranted to explore the therapeutic implications of targeting METTL14 and its downstream signaling pathways in clinical settings.
Comments (0)