Millets, ancient grains belonging to the Poaceae family, are often categorized as inferior coarse cereals. However, these grains are increasingly attracting global interest due to their superior nutritional profile and associated health benefits compared to more commonly consumed cereal staples. As a result, researchers are re-evaluating them as “super cereals.” Despite their potential, smallholder farmers, typically working with limited resources and minimal inputs, grow these grains primarily on marginal lands (Vetriventhan et al., 2020; Palanna et al., 2023a, 2023b). Among the minor millets, proso millet (Panicum miliaceum L.) stands out as an important crop, cultivated for food, feed, and fodder across Asia, particularly in India, Nepal, Pakistan, Myanmar, and Sri Lanka. Although small millets are known for their resilience to adverse environmental conditions, the emergence of banded sheath blight (Bsb), a disease caused by the pathogenic fungus Rhizoctonia solani, has become a major threat. This fungal infection has recently led to significant crop losses in many small millets, including proso millet (Patro et al., 2024).
The soil-borne pathogen R. solani (teleomorph: Thanatephorus cucumeris) is a notorious threat, known to infect over 100 plant species and potentially impacting a wide range of cultivated crops (Chahal et al., 2003; Ajayi-Oyetunde and Bradley, 2018; Patro et al., 2020). Isolates of R. solani are classified into 14 anastomosis groups (AG1 to AG13 and AGB1) based on their ability to undergo mycelial fusion, a trait that reflects their genetic similarities. These groups are further subdivided into 28 intergenic groups, characterized by differences in host range, pathogenicity, and genetic and biochemical properties (Carling et al., 2002; Singh et al., 2019). Given the global significance of R. solani and its intricate genetic diversity, extensive research is underway to explore the molecular mechanisms underlying its pathogenicity, employing a variety of omics approaches to deepen our understanding (Razali et al., 2021).
In nature, plants and pathogens engage in a constant battle, co-evolving as they adapt to one another’s strategies. Plants have developed sophisticated mechanisms to recognize and respond to pathogens, triggering defense systems that activate innate immunity to prevent invasion. The two primary defense mechanisms plants employ are PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006). However, pathogens have countered these defenses by evolving the ability to overcome host resistance through the secretion of key pathogenic components known as effectors (Stergiopoulos and de Wit, 2009; Sunani et al., 2024). These effector proteins are typically cysteine-rich, an amino acid containing sulfur that forms disulfide bridges, thus stabilizing their tertiary structures and making them more resistant to degradation (Ciuffetti et al., 2010; Khang et al., 2010). It is hypothesized that conserved motifs within fungal effectors, such as the Y/F/WxC, LFLAKHVLVXXP, and RXLR motifs, play crucial roles in effector uptake, suggesting that these motifs have specialized functions in pathogenesis (Liu et al., 2024).
Moreover, carbohydrate-active enzymes (CAZymes) are vital pathogenic factors secreted by plant pathogenic fungi, enabling them to breach plant cell walls and gain access to host tissues (Rytioja et al., 2014). In a previous secretome analysis of R. solani (RAP2) infecting barnyard millet (Echinochloa frumentacea), the top three predicted CAZymes were glycoside hydrolases, auxiliary activities, and glycosyltransferases. Additionally, a considerable number of secreted proteins from the virulence factor or pectin lyase fold superfamily were identified, which hydrolyze plant cell wall polymers to release saccharides. Notably, a comparison of the cutinase protein domains of R. solani strains infecting rice, maize, and soybean revealed a 28-bp deletion in the cutinase gene of the RAP2 strain. This deletion may play a critical role in enabling the pathogen to infect barnyard millet, potentially allowing it to bypass the host’s defense mechanisms (Patro et al., 2024).
Thus, pan-genome analysis offers valuable insights into the molecular basis of pathogenesis, host adaptability, and genetic diversity within species by distinguishing core from accessory genes. This approach also aids in identifying key genes involved in pathogenicity, host adaptation, and potential drug targets. To investigate the genetic composition of R. solani infecting proso millet, the current study aims to: (1) analyze the virulence and characterize R. solani isolates responsible for banded sheath blight (Bsb) disease in proso millet, and (2) employ genome sequencing and comparative analysis to explore the similarities and differences between R. solani strains infecting various crops.
Materials and methods Collection of disease samples, isolation, and purification of pathogenDuring the Kharif season of 2021, disease samples were collected from proso millet plants exhibiting symptoms of banded sheath blight. These samples, comprising infected leaves and leaf sheaths, were gathered from the Indian states of Odisha, Chhattisgarh, and Andhra Pradesh, as part of a survey (Table 1).
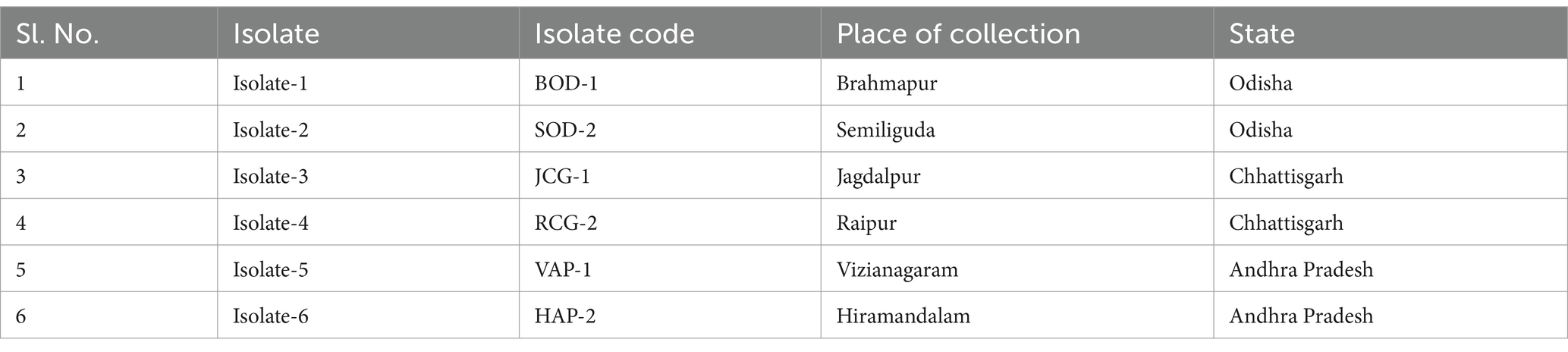
Table 1. Details of isolates gathered from various proso millet-growing regions across India for this study.
Proso millet leaves and leaf sheaths showing characteristic symptoms of banded sheath blight, collected from various locations were used for pathogen isolation on Potato Dextrose Agar (PDA) medium. Leaf bits measuring approximately 4 mm2, including both healthy and infected portions, were excised and surface sterilized using 1% sodium hypochlorite for 1 min. This was followed by three rinses with sterilized distilled water to ensure complete removal of the disinfectant. The sterilized leaf segments were then placed between sterile blotting sheets to remove excess moisture before being aseptically transferred to PDA plates (Ramesh et al., 2021; Sunani et al., 2025). The plates were incubated at 27 ± 1°C, and observations for mycelial growth were made at 24-h intervals. After 2 days, mycelial bits from the diseased leaf segments were aseptically transferred onto glass slides for microscopic examination. Morphological characteristics, including hyphal width, branching angle, and septation, were assessed to confirm the pathogen’s identity. The confirmed pathogen cultures were subsequently sub-cultured on PDA for further study (Singh et al., 2018).
Pathogenicity and virulence analysisThe pathogenicity of the test isolates was assessed by inoculating one-month-old plants of the susceptible variety, Nilavoor local. Seedlings were cultivated in plastic pots measuring 30 cm in diameter and 20 cm in depth, containing a sterilized soil and vermicompost mixture in a 2:1 ratio. The leaf sheath was inoculated following the method outlined by Shekhar and Kumar (2012). Each pot contained two plants, with three replications maintained for each treatment. Control plants, which were not inoculated with the pathogen, were also included. Disease responses and the percent disease index were assessed 25 days’ post-inoculation using the standard assessment scale described by Palanna and Das (2022).
Cultural and morphology analysisCultural characteristics were recorded when the hyphae reached the edge of the Petri dishes, focusing on observations of colony color, growth pattern, colony texture, and hyphal width. Morphological diversity among the isolates was assessed based on hyphal characteristics and sclerotial features, including their color, texture, size, and formation patterns, as well as the arrangement of sclerotia. Mycelial masses from a 10-day-old culture were mounted and photographed using an Olympus BX 51 microscope equipped with a Progres 2.7 (Jenoptik, United States) digital camera for microscopic examination (Ram et al., 2024). Sclerotial characteristics were recorded by analyzing an average of 25 sclerotia for each isolate.
DNA extraction, PCR amplification and phylogenetic analysisCultures incubated for 5 days at 27 ± 1°C on potato dextrose broth (PDB) and inoculated with mycelial discs were used for DNA extraction following the CTAB method (Murray and Thompson, 1980). DNA concentration was measured using a Nanodrop spectrophotometer (JENWAY Genova Nano Ver.1.58.3) at the 260/280 nm spectrum. The ITS region of rDNA from all six isolates was amplified using the primer pair ITS-1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS-4 (5′-TCCTCCGCTTATTGATATGC-3′) with an annealing temperature of 57°C (Patro et al., 2024). The amplified PCR products were sequenced. The obtained sequences were aligned using Clustal W. Additional ITS-5.8S rDNA sequences of R. solani isolates representing different anastomosis groups from the NCBI GenBank were included for phylogenetic analysis. A phylogram illustrating the relationships among R. solani isolates was constructed using the neighbor-joining method (Patil et al., 2024; Sandilya et al., 2022).
Genome sequencing, assembly, gene prediction, and annotationDNA libraries were prepared using the TruSeq DNA Nano LP kit and quantified with a Qubit 4 Fluorometer. Sequencing was performed on the Illumina NovaSeq 6,000 platform, generating 154 bp paired-end reads. The quality of the raw reads was assessed using FastQC v. 0.11.9 (Andrews, 2019). Subsequently, adapter sequences were removed, and quality filtering was applied using default parameters. Additionally, the first 10 bases of the 5′ end of each paired-end read were trimmed using Trim Galore v.0.6.5 (Felix, 2022).
The adapter-free, quality-filtered reads were assembled de novo with default settings using SPAdes v.3.15.5 (Bankevich et al., 2012). Genome-guided assembly correction and scaffolding were performed on the finished assembly using the R. solani AG-1 IA strain RefSeq genome (GCF_016906535.1) as a reference, with RagTag v.2.1.0 (Alonge et al., 2022). To eliminate repetitive contigs, the assembly scaffold was further refined using Funannotate v. 1.8.7 (Palmer and Stajich, 2020), with contigs shorter than 500 bp removed to generate the final genome. The adapter-free, quality-filtered reads were then mapped back to the finished genome using the bwa-mem v.0.7.18 (Heng, 2013) aligner with default settings, to evaluate the read utilization in the assembly process. Genome assembly quality was assessed using QUAST v.5.2.0 (Gurevich et al., 2013), and genome completeness was evaluated with CEGMA (Parra et al., 2007) on the gVolante (Nishimura et al., 2017) web server using the default settings and the CEGMA eukaryotic ortholog set for non-vertebrate gene prediction.
Gene prediction was performed using the “predict” command of the Funannotate v.1.8.7 pipeline, with RNA sequences from the R. solani AG-1 IA strain RefSeq (GCF_016906535.1) genome used as transcript evidence. Predicted protein sequences were annotated by creating a local BLAST database of UniProt proteins (The UniProt Consortium, 2023), followed by homology searches using Protein BLAST (Altschul et al., 1990; Camacho et al., 2008).
Repetitive genomic elements of the VAP-1 genomeThe repetitive landscape of the VAP-1 genome was characterized through the identification and annotation of repetitive elements. De novo prediction of VAP-1 specific repetitive sequences was performed using RepeatModeler v.2.0.3 (Flynn et al., 2020), and sequences with hits to known elements were annotated using the Repbase release v.20181026 (Bao et al., 2015). RepeatMasker v.4.1.3-p2 (Tarailo-Graovac and Chen, 2009) was employed to mask and annotate simple repeats, curated fungal repeats from the Repbase database, and genome-specific repeats. Additionally, the VAP-1 genome was scanned for simple sequence repeats, which were classified using the MegaSSR online server (Morad et al., 2023).
Pathway and Gene Ontology analysisThe protein sequences of the VAP-1 assembly were subjected to analysis using BlastKOALA (Kanehisa et al., 2016) to determine their Kegg Orthology (KO) identifiers. Pathway annotations were then carried out through KEGG Mapper (Kanehisa and Sato, 2020; Kanehisa et al., 2022). To further investigate the functional implications, the UniprotR package v.2.3.0 (Soudy et al., 2020) was employed to query Gene Ontology (GO) terms, identifying the top 10 enriched terms associated with biological processes, molecular functions, and subcellular localization.
Secretome analysisTo predict the fungal secretome of the VAP-1 genome, a standardized workflow was established, mirroring the methodology used in a previous study for the in silico secretome prediction of the ascomycete fungus Parascedosporium putredinis NO1 (Scott et al., 2023). For subcellular localization prediction, the protein sequences of the VAP-1 genome were input into the DeepLoc v. 2.1 server (Ødum et al., 2024). Subsequently, secretion prediction was performed using the SignalP v. 6.0 (Teufel et al., 2022) and TargetP v. 2.0 (Armenteros et al., 2019) servers. The resulting localization and secretory protein sequences were then cross-referenced to remove redundancies, retaining only unique proteins. These unique proteins were further analyzed using DeepTMHMM v. 1.0 (Hallgren et al., 2022) to identify potential transmembrane proteins. Among these, proteins predicted to possess alpha-helical or beta-barrel structures as transmembrane proteins were considered as part of the secretome of the VAP-1 genome. Superfamily annotations were derived by screening the secretome proteins against the InterPro protein signature databases (Paysan-Lafosse et al., 2022). Carbohydrate-active enzymes (CAZymes) in the VAP-1 genome were annotated using the dbCAN3 web server (Jinfang et al., 2023), employing the HMMER tool for CAZyme family identification through a search against the dbCAN CAZyme domain HMM database. Additionally, CAZyme subfamily annotations were obtained via an HMMER search against the dbCAN-sub HMM database, which categorizes CAZyme subfamilies.
Comparative genomics analysisThe genome of the highly virulent VAP-1 isolate of R. solani from proso millet, which belongs to the Poaceae family, was compared to the genomes of other Poaceae species, such as rice. Additionally, to explore broader genetic relationships, the VAP-1 genome was also compared with those of R. solani strains infecting non-Poaceae plants, including sugar beet and tobacco. The genome sequences for R. solani strains infecting rice (GCA_015342415.1), sugar beet (GCA_905219575.1), and tobacco (GCA_905219615.1) were retrieved from the NCBI Genomes database (Sayers et al., 2021). These genomic datasets were uploaded to the MOSGA web server (Martin et al., 2020, 2021) to assess genome completeness and to calculate the average nucleotide identity (ANI) between the species. To further investigate functional similarities, the protein sequences of the four R. solani genomes were analyzed through the OrthoVenn3 web server (Sun et al., 2023), which identified orthologous gene clusters and provided insights into their functional relevance across the genomes.
Variation and evolution of secretome proteinsThe secretome protein repertoire of R. solani (VAP-1), comprising 124 proteins, was analyzed with R. solani genome’s proteins from rice, sugar beet, and tobacco. This analysis was conducted through a homologous search using BLASTP v.2.10.0+ with the 124 secretome proteins of R. solani (VAP-1) as queries against the complete protein sequences of R. solani from rice, sugar beet, and tobacco. Homologous sequences with more than 70% identity were retained for further analysis. The homologous secretome proteins from R. solani infecting proso millet (VAP-1), along with those from rice, sugar beet, and tobacco, were submitted individually to the OrthoVenn3 web server (Sun et al., 2023) for gene family contraction and expansion analysis using CAFE (Mendes et al., 2020). A comparative analysis of the secretome proteins annotated with InterPro superfamily signatures, along with their homologous proteins from R. solani genomes infecting rice, sugar beet, and tobacco, was performed. This analysis focused on identifying variations in the conserved sequences of the proteins. Multiple sequence alignments of Major Facilitator Superfamily (MFS), transglycosidases, P-loop containing nucleoside triphosphate hydrolases, and galactose oxidases were conducted using the Clustal Omega web server (Sievers and Higgins, 2018).
Results Disease symptoms, and pathogen isolationBsb symptoms initially appear as small, irregularly shaped white lesions on the lower leaf sheath, located between the soil line and the leaf blade. Over time, these lesions expand to the upper leaf sheaths, forming 2–3 cm long oval to irregular lesions that transition from light grey to dark brown. Eventually, the lesion centers become greyish white to straw-colored, bordered by narrow reddish-brown margins. These margins form a series of brown lines along the leaf sheaths, creating a distinct banded appearance. As the disease progresses, the lesions merge, enlarge, and extend upward, ultimately causing blighting of the leaf sheath and leaf blade (Figure 1). Bsb diseases samples were obtained from infected proso millet plants across various regions of Indian states and were purified using the hyphal tip method, following standard isolation techniques. Six R. solani isolates were recovered, each assigned a unique identity code (Table 1).

Figure 1. Symptoms of banded sheath blight on proso millet include: (a) initial appearance of small lesions on the leaf sheath. (b) Development of elongated, oval-shaped lesions on the stem. (c) A characteristic banded appearance on the leaves, marked by straw-colored lesions with distinct brown margins. (d) Advanced stages affecting the panicle, leading to grain discoloration and drying.
Morphology and cultural characteristicsOn PDA medium, the fungus formed yellow, cottony, fluffy colonies with both aerial and surface mycelia. The hyphal width ranged from 5.02 ± 0.20 μm to 7.42 ± 0.17 μm. Significant variation was observed among the isolates in their sclerotial characteristics. Sclerotia were formed either aerially or on the surface, with their color ranging from light brown to dark brown. The sclerotial diameter varied from 0.93 ± 0.05 mm to 1.79 ± 0.01 mm. The texture of the sclerotia was either rough or smooth, and their distribution was central, sub-central, or in a peripheral ring. Among the isolates, RCG-02 produced the highest number of sclerotia (32), while JCG-01 produced the fewest (13) (Supplementary Table 1) (see Figure 2).

Figure 2. Morphological characteristics of R. solani isolates. (I) Colony morphology of R. solani isolates (a) BOD-1, (b) SOD-2, (c) JCG-1 (d) RCG-2 (e) VAP-1 (f) HAP-2. (II) Microscopic images (400×) depicting the hyphal characteristics of R. solani. (a) Right angle branching. (b) Constriction at point of origin. (c) Hyphal fusion. (d) Barrel shaped monilioid cells.
Pathogenicity and virulence analysisThe PDA plugs, containing actively growing mycelium of different isolates of R. solani, were inoculated on susceptible proso millet cultivar, Nilavoor local. All six isolates produced typical symptoms on leaf sheath 4 to 8 days’ post-inoculation. Using a standard scale (Palanna and Das, 2022), the virulence of the isolates was ascertained based on the percentage disease index (PDI). Nilavoor local showed typical lesions with ranged values of PDI from 22.60 to 65.50%. VAP-1 was the most virulent among the isolates, with a disease incidence of 65.65%, followed by HAP-2 (58.70%), RCG-2 (58.50%), BOD-1 (47.75%), and SOD-2 (41.54%), while JCG-1 had the lowest incidence of disease (22.60%).
Molecular analysis to confirm the pathogenPCR amplification of the ITS region resulted in a fragment size of approximately 600 bp in length. The gene fragments were purified, and sequenced. Further, the gene sequences were submitted at NCBI GenBank. A phylogenetic tree was generated using the different gene sequences with various AGs and the ITS sequences from six R. solani isolates. All the six isolates showed highest similarity per cent with the AG-1 1A group of R. solani which confirms that all the six isolates belong to the anastomosis group, AG-1 1A subgroup (Figure 3).
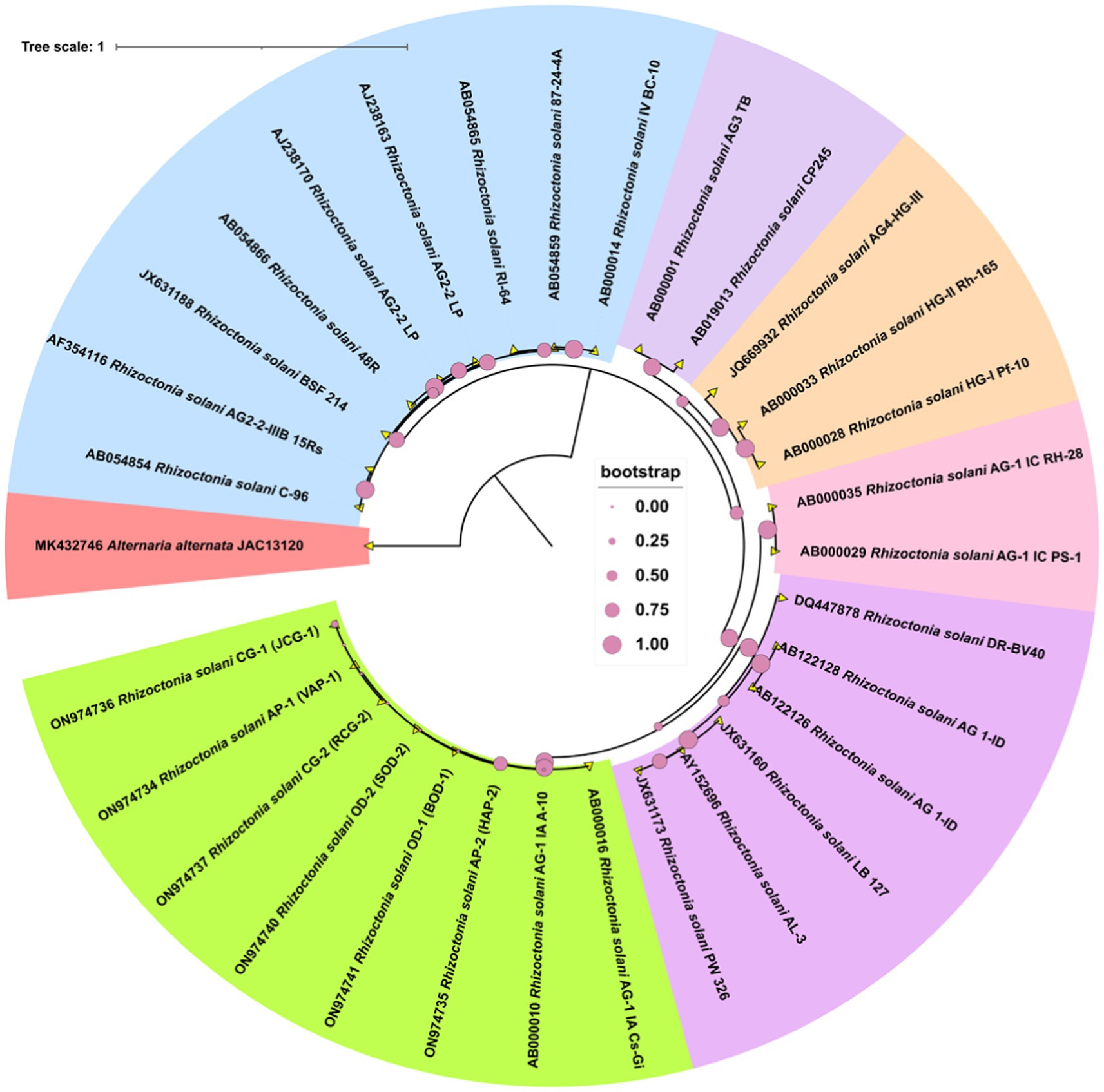
Figure 3. The phylogram generated from ITS sequences using the neighbor-joining method illustrates the genetic relationships among R. solani isolates. Bootstrap values, calculated from 1,000 replicates to produce a majority consensus tree, are indicated on the branches. The strain Alternaria alternata (JAC13120) was used as an outgroup for the analysis.
Genome sequencing, assembly, gene prediction, and functional annotationPost virulence studies, the VAP-1 strain, identified as the most virulent, was selected for whole genome sequencing using the Illumina platform. The raw sequencing data consisted of 7.2 million 154 bp paired-end reads (Supplementary Table 2), with 96% of the reads having a Phred score greater than 30 (Figure 4A). After quality filtering, the library retained approximately 99.98% of the initial reads (Supplementary Table 2), with 98% of the reads having a Phred score greater than 30 (Figure 4B).
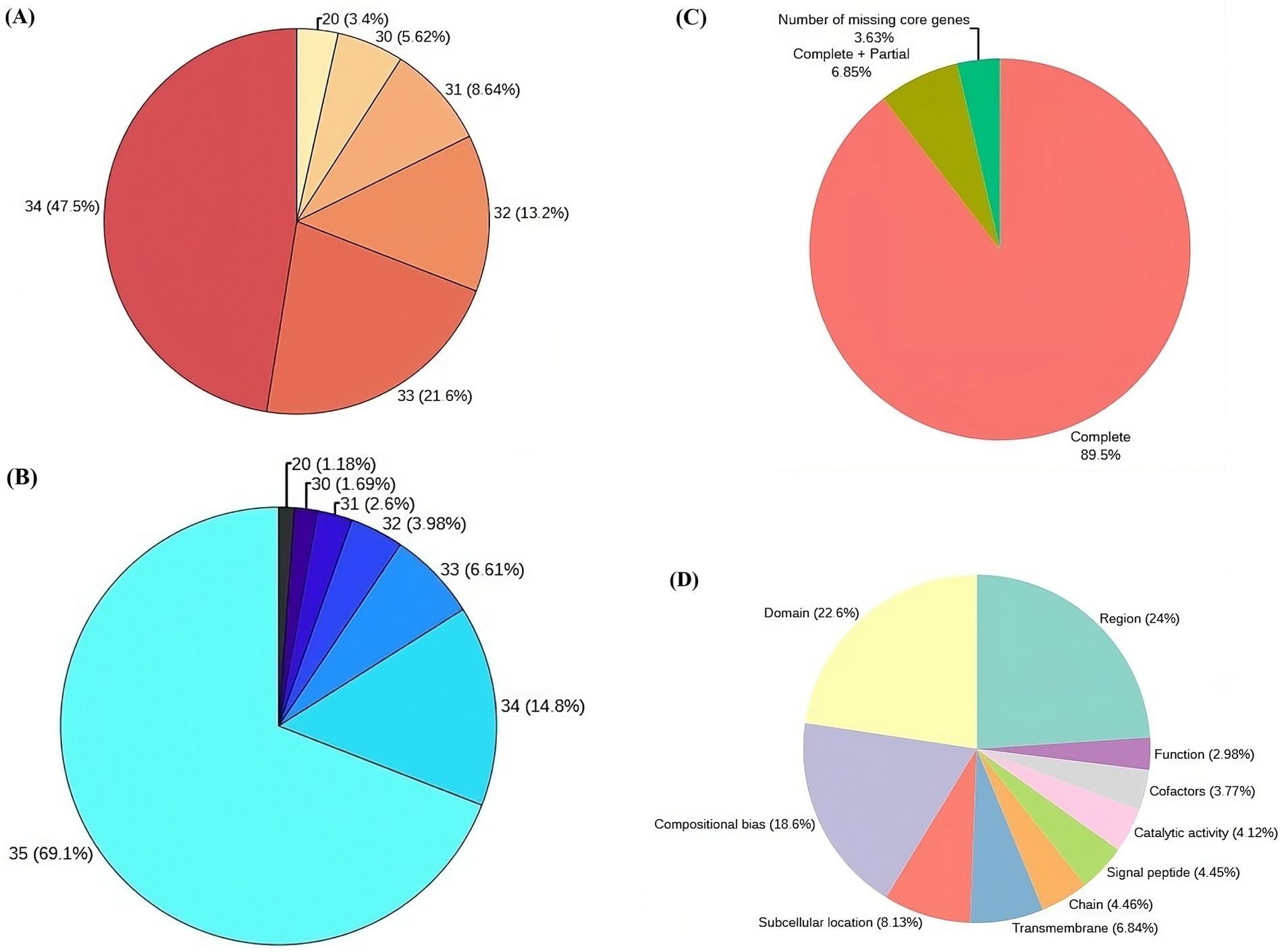
Figure 4. Genome sequencing, assembly, and protein annotation statistics. (A) Raw read statistics, with each segment annotated by Phred score and the corresponding percentage of reads in the library. (B) Quality-filtered read statistics, including Phred scores and the percentage of reads in the library. (C) CEGMA genome assembly completeness statistics (D) Top 10 sequence annotation features of proteins associated with UniProt.
Following quality filtration, the de novo assembly of the reads resulted in a genome comprising 6,096 contigs, totaling approximately 47.12 Mb. Read utilization analysis indicated that 99.55% of the quality-filtered reads successfully mapped back to this assembled genome. The largest contig accounted for 8.3% of the total genome length, with N50 and N90 contig lengths of 2,122,470 bp and 2,398 bp, respectively. Additionally, the assembly’s L50 and L90 values were 8 and 1,207, respectively (Supplementary Table 3). When the assembly was evaluated against the eukaryotic ortholog dataset of 248 core non-vertebrate genes, it demonstrated 89.5% completeness, with 6.85% of genes partially complete and 3.63% missing (Figure 4C).
Gene prediction of the VAP-1 genome identified 9,321 protein sequences, which were subsequently annotated by comparing them to the 22,082 R. solani species-specific proteins obtained from the UniProt database. A homology search using BLASTP v.2.10.0+ revealed that approximately 81% of the 9,321 proteins exhibited more than 90% homology with R. solani specific protein sequences (Supplementary Table 4). Further analysis of the VAP-1 genome’s protein sequences annotations showed that the top three UniProt sequence annotation features of the sequences were region, domain, and compositional bias, comprising 24, 22.6, and 18.6% of the proteins, respectively (Figure 4D).
Repetitive genomic elements of the VAP-1 genomeThe repetitive landscape of the VAP-1 genome constitutes approximately 17.62% of its total sequence, with 97.8% composed of interspersed repeats, followed by minor contributions from simple repeats, low-complexity regions, and small RNA elements (Figure 5A). Interspersed repeats are predominantly composed of retroelements (72.35%), DNA transposons (5.47%), and unclassified repeats (22.18%). The retroelements are largely made up of LTR elements (97.29%) and LINEs (2.71%). Among the LTR elements, Ty3-retrotransposons account for 95.68%, while Ty1/Copia elements represent 4.32%. The DNA transposons are primarily composed of MULE-MuDR (72.55%) and Tc1-IS630-Pogo (27.45%) (Figure 5B). The most common simple sequence repeat (SSR) classes in the VAP-1 genome are di-, penta-, and hexanucleotide repeats (Figure 5C). The dominant dinucleotide SSRs are “AA” and “GG” (Figure 5D), while the penta-and hexanucleotide SSRs are “TTTTT” (Figure 5E) and “TTTTTT” (Figure 5F).
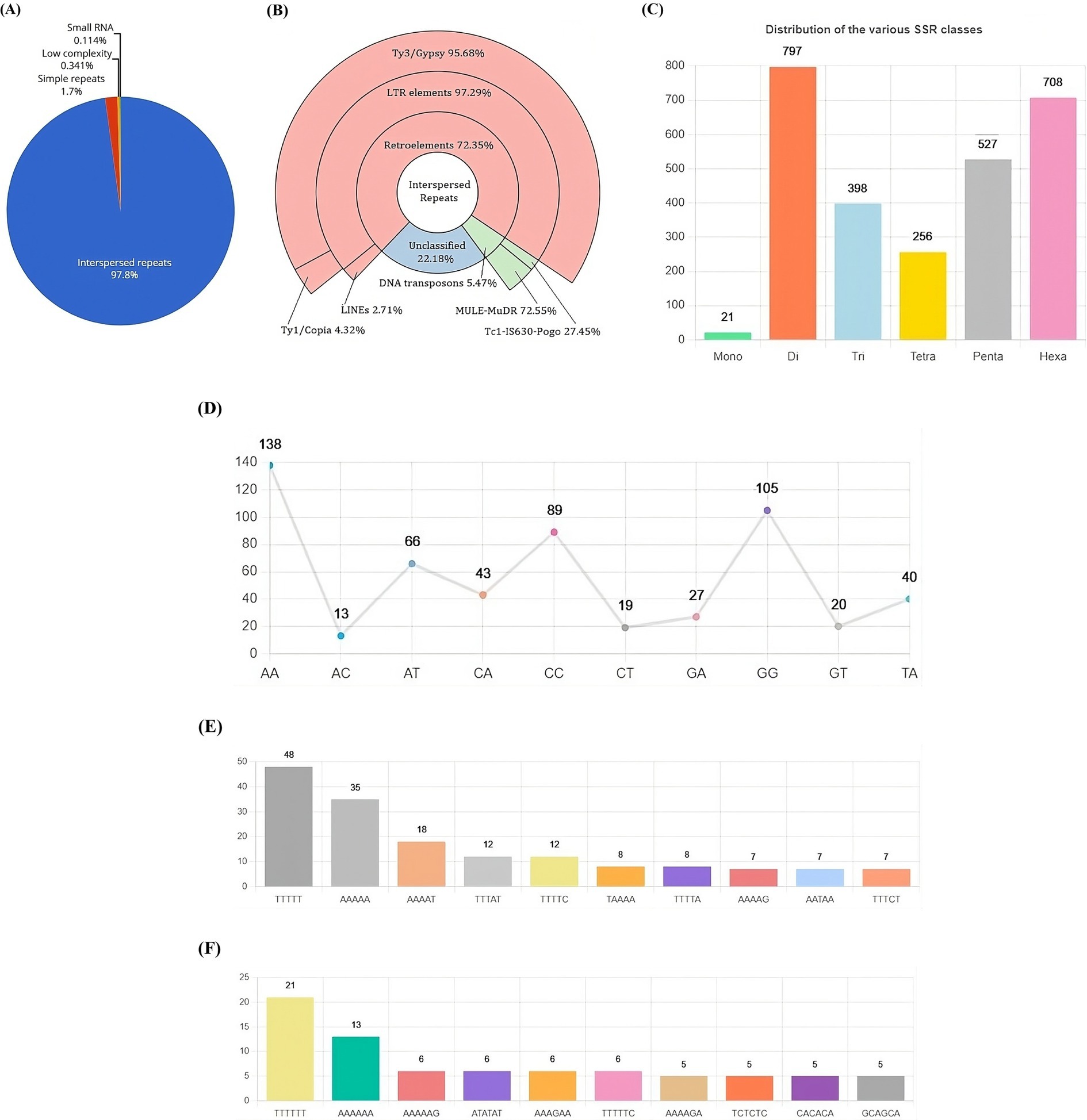
Figure 5. Repetitive DNA landscape of the R. solani VAP-1 isolate genome. (A) Composition of repetitive DNA. (B) Classification of interspersed repeats. (C) Distribution of SSR classes. (D) Proportion of di-nucleotide SSRs. (E,F) Distribution of top 10 penta-and hexa-nucleotide SSRs.
Pathway and Gene Ontology analysisThe top three enriched KEGG pathway maps in the “Metabolism” category were “Carbohydrate metabolism,” “Amino acid metabolism,” and “Metabolism of cofactors and vitamins.” In the “Genetic Information Processing” category, the most enriched pathways included “Translation,” “Folding, sorting, and degradation,” and “Replication and repair.” The “Environmental Information Processing” category was dominated by the “Signal transduction” and “Membrane transport” pathways, while the “Cellular processes” category showed enrichment in “Transport and catabolism,” “Cell growth and death,” and “Cellular community-eukaryotes” (Figure 6A). Gene Ontology analysis revealed that the terms “proteolysis,” “membrane,” and “ATP binding” exhibited the highest gene enrichment within the “biological process,” “cellular component,” and “molecular function” domains, respectively (Figure 6B).
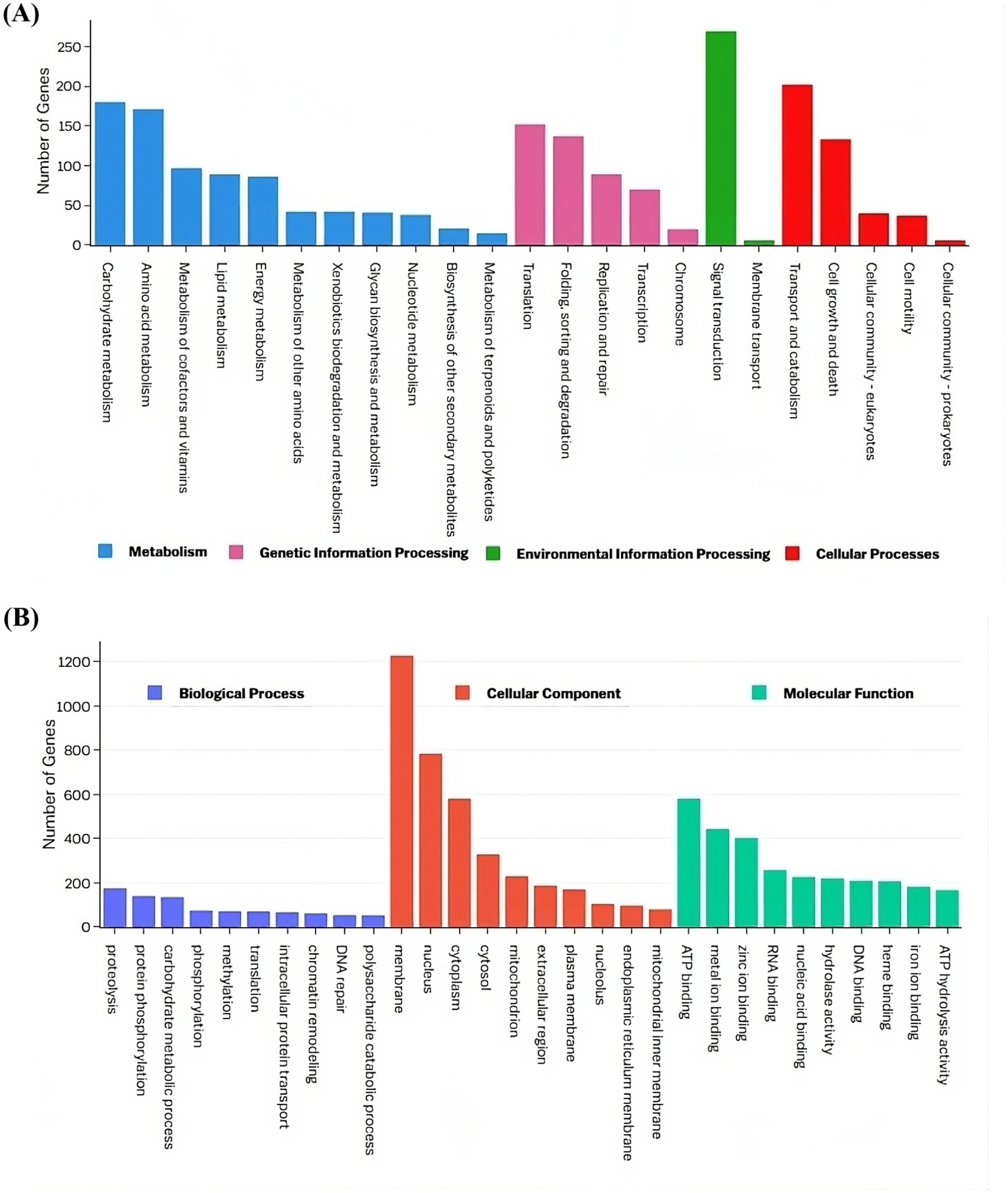
Figure 6. Functional annotation of the VAP-1 genome proteins infecting proso millet. (A) The KEGG annotation chart, with legends indicating the Level 1 hierarchy, where the bars represent the number of genes involved in the Level 2 pathways that fall under each Level 1 category. (B) The GO functional annotation classification statistics plot, where the y-axis represents the number of genes, and the x-axis displays a set of ontology terms corresponding to the Gene Ontology domains: molecular function, cellular components, and biological process.
Secretome analysisSecretome prediction began with the identification of subcellular localization and secretion proteins, resulting in the prediction of 933 proteins as localization proteins, and 854 and 688 proteins as secretory by the TargetP and SignalP tools, respectively (Figure 7A). The majority of these proteins (53%) were predicted to be both localization and secretory proteins (Figure 7B). Subsequently, 1,146 unique proteins were analyzed for the presence of alpha-helical or beta-barrel structures. Of these, 124 proteins were identified by DeepTMHMM as transmembrane proteins, which were then classified as part of the secretome of the VAP-1 genome. Further analysis using the EffectorP tool predicted 20 of these secretome proteins to contain signal peptides indicative of secretion. Superfamily annotations of the secretome proteins revealed that the top five categories included the MFS general substrate transporter, (Trans) glycosidases, P-loop containing nucleoside triphosphate hydrolases, SGNH hydrolase families and Galactose oxidase, central domain (Figure 7C). CAZyme classification identified the top five predicted enzymes as belonging to the Glycoside Hydrolases class, followed by Auxiliary Activities, Polysaccharide Lyases, Glycosyltransferases, and Carbohydrate Esterases classes (Figure 7D).
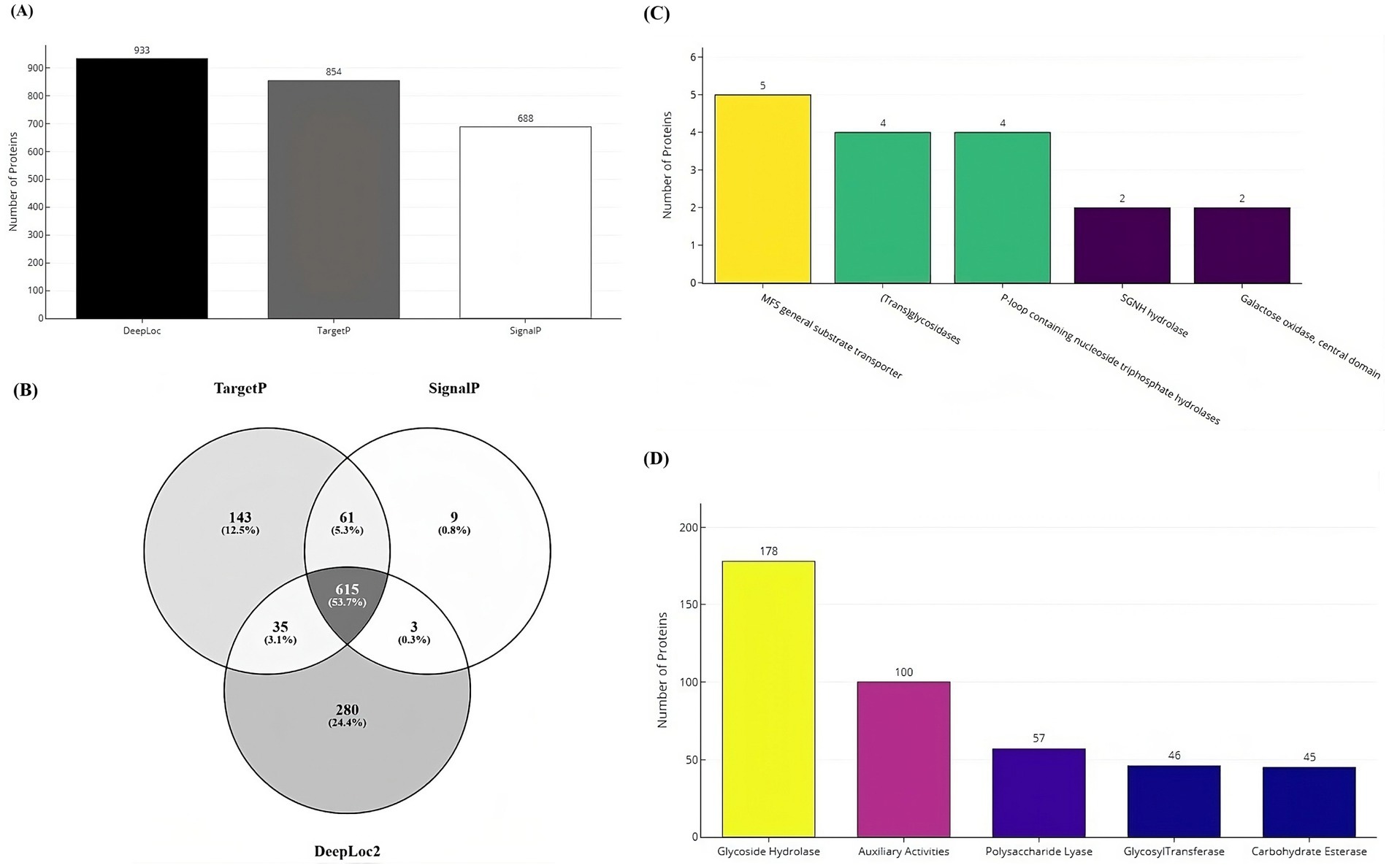
Figure 7. Secretome and CAZyme analysis of the VAP-1 genome. (A) Number of localization and secretory proteins predicted by the DeepLoc, TargetP, and SignalP tools. (B) Number of proteins shared across the DeepLoc, TargetP, and SignalP predictions. (C) SUPERFAMILY annotations of secretome proteins (D) Distribution of CAZyme classes predicted in the VAP-1 genome.
Comparative genomics analysisThe gene content analysis revealed that the VAP-1 R. solani genome contained over 80% of complete BUSCOs from the 260 BUSCOs in the Eukaryota dataset. This was followed by a small proportion of fragmented and duplicated BUSCOs, with approximately 10% of BUSCOs missing. Among the genomes compared, the R. solani genome isolated from rice exhibited slightly superior BUSCO completeness compared to the VAP-1 genome, while R. solani genomes from sugar beet and tobacco showed fewer complete BUSCOs than VAP-1. In the comparison with the 260 BUSCOs from the Eukaryota dataset, the VAP-1 genome was found to be missing three orthologs, which were absent in all other R. solani genomes. Notably, the VAP-1 genome lacked one BUSCO, also absent in the tobacco and rice R. solani genomes, and another BUSCO that was missing in both VAP-1 and rice genomes. Three specific BUSCOs were missing only from the VAP-1 genome (Figure 8A).
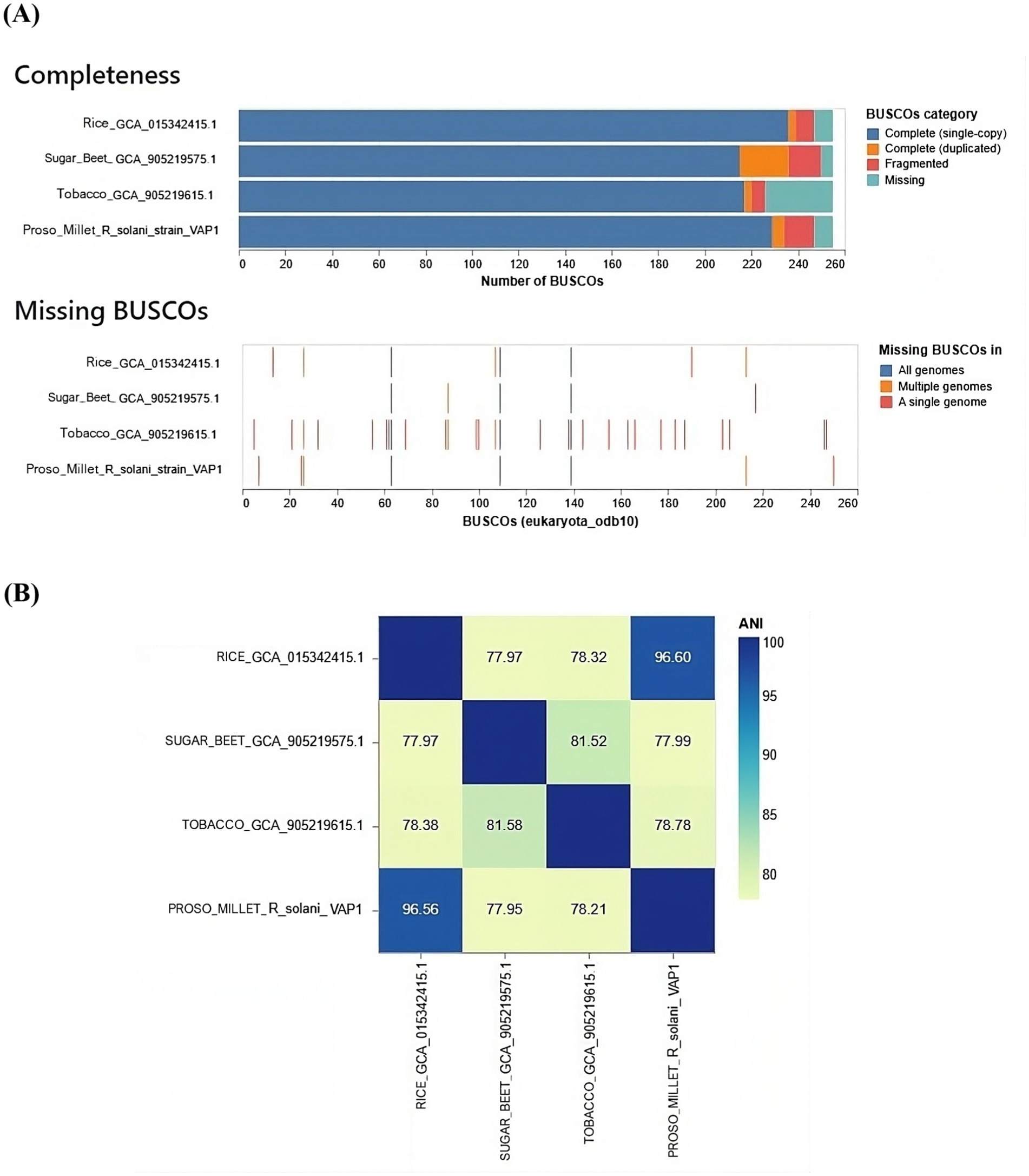
Figure 8. Comparative genomic analysis of R. solani genomes. (A) The assembly completeness of the R. solani genomes evaluated using the Eukaryota dataset of 260 BUSCOs, with the analysis quantifying both complete and missing BUSCOs. (B) The average nucleotide identity (ANI) as a measure of the genomic similarity among the R. solani genomes.
The VAP-1 genome infecting proso millet and the R. solani genome infecting rice exhibited ANI values greater than 96%, suggesting a close species-level relationship. In contrast, the ANI values between the VAP-1 genome and the R. solani genomes infecting sugar beet and tobacco were below 96%, indicating a greater degree of genomic divergence (Figure 8B).
OrthoVenn3 analysis of VAP-1 protein sequences, compared with those from R. solani genomes infecting rice, sugar beet, and tobacco, identified 11 shared homologous gene clusters and 4 unique clusters across all species. A single large homologous gene cluster containing 4,028 proteins was common to all fungal genomes, while a unique cluster of 105 proteins was specific to the VAP-1 genome (Figure 9A). Functional Gene Ontology (GO) analysis of the 4,028 shared proteins revealed that the top three GO terms in the Biological Process domain were GO:0008150 (Biological process), GO:0008152 (Metabolic process), and GO:0044237 (Cellular metabolic process). In the Molecular Function domain, the top three GO terms were GO:0016491 (Oxidoreductase activity), GO:0016740 (Transferase activity), and GO:0016787 (Hydrolase activity). The top three GO terms in the Cellular Component domain were GO:0016020 (Membrane), GO:0005575 (Cellular component), and GO:0044464 (Cell part) (Figure 9B). The unique cluster of 105 proteins specific to the VAP-1 genome was enriched with five significantly associated GO terms, all within the Biological Process domain. These terms included GO:0033609 (oxalate metabolic process), GO:0009405 (pathogenesis), GO:0000272 (polysaccharide catabolic process), GO:0016052 (carbohydrate catabolic process), and GO:0009853 (photorespiration) (Table 2).
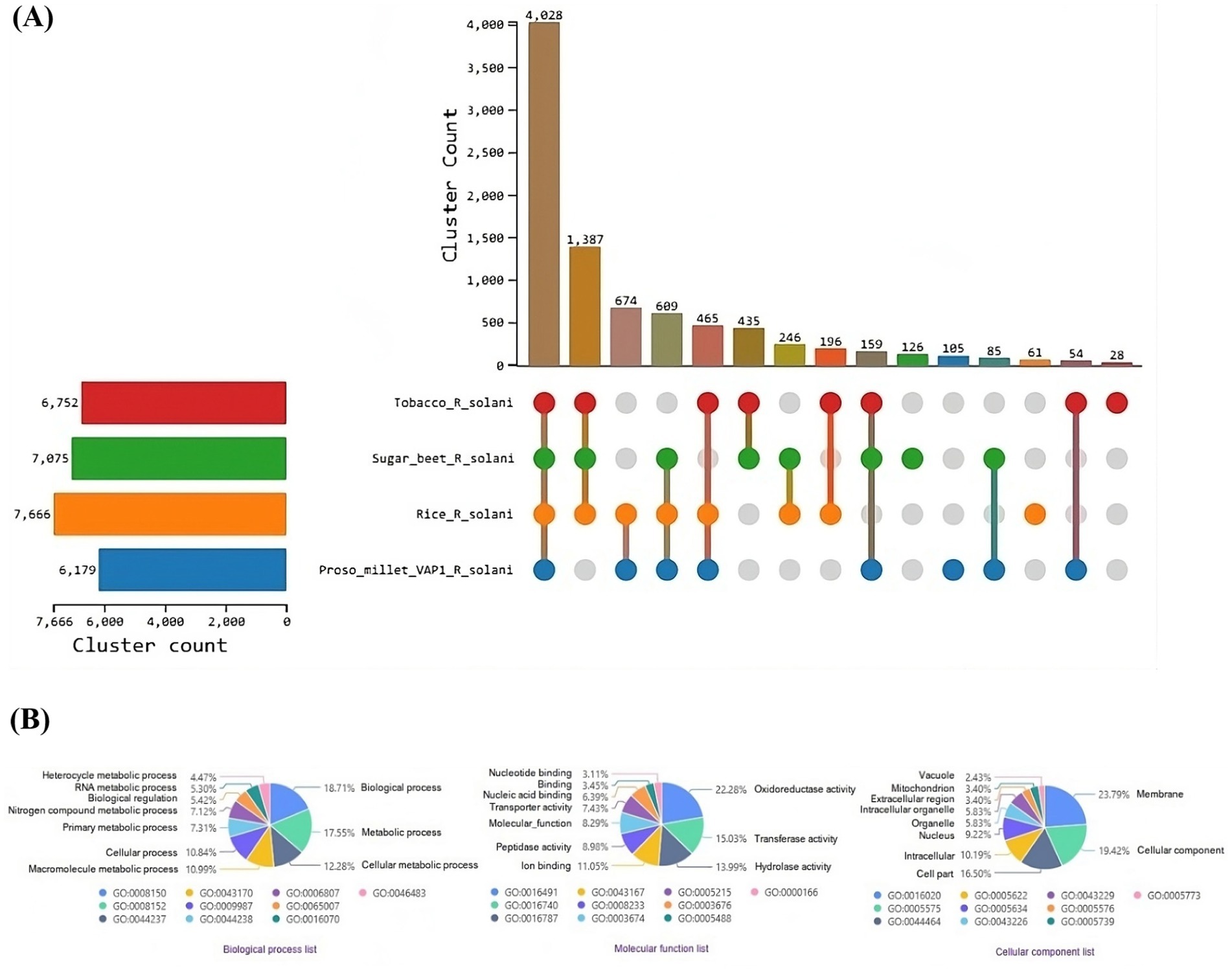
Figure 9. Comparative genomic analysis of protein sequences from R. solani genomes. (A) UpSet plot illustrating the distribution of orthologous protein clusters across the different R. solani genomes. (B) Enriched Gene Ontology (GO) terms of the shared orthologous protein clusters present in the R. solani genomes.
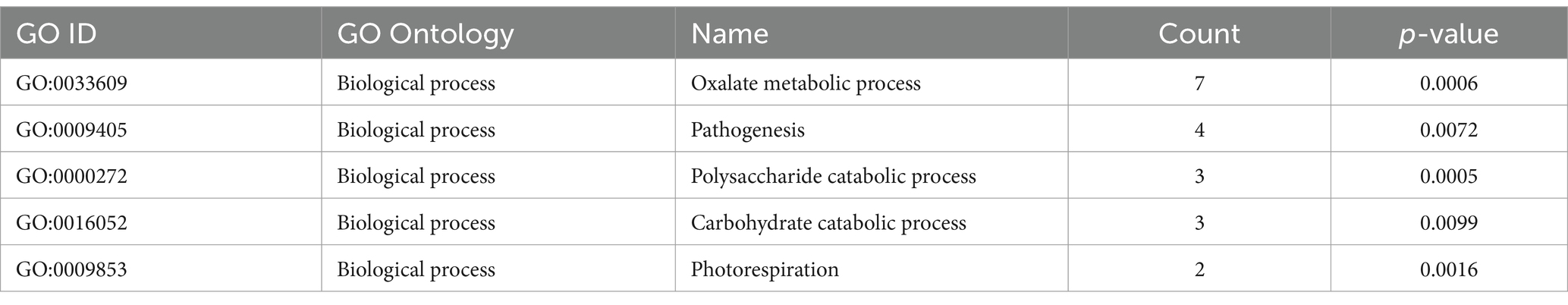
Table 2. OrthoVenn3 GO enrichment analysis of proteins clustered specifically to VAP-1 genome.
Variation and evolution of secretome proteinsOrthologous analysis of the secretome protein sequences from R. solani infecting proso millet, rice, sugar beet, and tobacco revealed that the secretomes of R. solani infecting sugar beet and tobacco have diverged into a distinct lineage, whereas the secretomes of R. solani infecting proso millet and rice form a separate, closely related lineage. Expansion and contraction analysis indicated that two gene families have contracted in R. solani infecting rice, while a single gene family has contracted in R. solani infecting sugar beet (Supplementary Figure S1A). A major facilitator superfamily protein, characterized as a transmembrane 9 superfamily protein from the VAP-1 genome, which shares greater than 70% identity with proteins from R. solani genomes of rice, sugar beet, and tobacco, was analyzed for variations. Multiple sequence alignment of these proteins revealed the presence of variable numbers of base deletions in R. solani infecting proso millet, rice, and sugar beet at different locations when compared to the tobacco isolate (Supplementary Figure S1B). Similarly, multiple sequence alignment of a single (Trans) glycosidases superfamily enzyme, specifically a Glycoside hydrolase family 10 protein, from R. solani infecting sugar beet, proso millet, and rice, revealed a single base mutation and consecutive two-base deletions in R. solani infecting proso millet and sugar beet when compared to rice isolate. The protein from R. solani infecting tobacco exhibited less than 70% identity and was therefore excluded from the analysis (Supplementary Figure S1C). Additionally, multiple sequence alignment of P-loop containing nucleoside triphosphate hydrolases, characterized as ATP-binding cassette subfamily B proteins from all R. solani genomes, revealed a 21-base deletion in R. solani infecting proso millet, located in a highly variable region surrounding the conserved region of the protein (Supplementary Figure S2A). Similarly, multiple sequence alignment of galactose oxidase proteins from R. solani infecting tobacco, proso millet, and rice revealed the presence of a 3-base deletion in the RAX2 domain in R. solani infecting tobacco and rice, compared to proso millet isolate (Supplementary Figure S2B).
DiscussionThe present study provides valuable insights into the pathological and molecular characteristics of R. solani isolates associated with Bsb in proso millet. Pathogenicity assays revealed significant variation in virulence among the R. solani isolates, with PDI values ranging from 22.60 to 65.50%. VAP-1 was identified as the most virulent isolate, with the highest disease incidence (65.65%), while JCG-1 exhibited the lowest virulence (22.60%). This variability underscores the significant intraspecific variability in fungal pathogens including R. solani, often associated to their complex genetic makeup and adaptability (Jeevan et al., 2023; Palanna et al., 2023a, 2023b). The findings emphasize the importance of region-specific screening of proso millet cultivars for resistance and the development of tailored fungicidal management strategies.
Molecular characterization further confirmed the identity of the isolates. PCR amplification, sequencing, and phylogenetic analysis of the ITS region identified all six isolates as belonging to the AG-1 1A subgroup of R. solani. This result aligns with previous studies indicating AG-1 1A as the primary anastomosis group associated with sheath blight in cereal crops (Liu et al., 2024; Patro et al., 2024). Given its high virulence, VAP-1 was selected for whole-genome sequencing to provide deeper insights into the genetic factors driving its pathogenicity.
In this study, we identified the most virulent R. solani strain (VAP-1) infecting proso millet and successfully sequenced, assembled, predicted, and annotated its functional genes. This work advances our understanding of the host’s genetic composition and offers valuable insights into the host-pathogen interactions between R. solani and proso millet. Sequencing on the Illumina platform generated high-quality paired-end reads, with 96% of the reads having a Phred score greater than 30. After applying quality filtration to remove adapter contamination, approximately 99.98% of the reads were retained for the genome assembly process. De novo assembly utilized 99.55% of the reads to assemble the VAP-1 genome, resulting in a total genome size of 47.12 Mb composed of 6,096 contigs. The largest contig is 3,918,178 bp in length, representing over 8% of the genome, while the N50 contig length is 2,122,470 bp, corresponding to the 8th largest contig (L50) in the assembly. The remaining 40% of the genome is composed of 1,199 contigs of varying sizes, with the N90 contig length measuring 2,398 bp.
The model reference strain (GCF_016906535.1) in the NCBI RefSeq database has a genome size of 40.85 megabases, consisting of 17 contigs. This is smaller compared to the VAP-1 genome in terms of both genome size and the number of contigs. The observed variation could be attributed to several factors, with the most significant being the repetitive content in phytopathogens, which may drive retrotransposon proliferation and genome size expansion (Spanu et al., 2010). Additionally, structural variations such as insertions, deletions, duplications, and inversions (Hartmann, 2021), as well as evolutionary adaptations to specific ecological niches (Naranjo-Ortiz and Gabaldón, 2019), may also contribute to the differences in genome characteristics.
Retrotransposons are known to play a critical role in fungal pathogenicity by promoting genome plasticity, regulating virulence factor expression, facilitating host adaptation, and conferring resistance to stress (Razali et al., 2019). In the VAP-1 genome, the majority of the repetitive content is composed of retrotransposons. Ty3-retrotransposons account for approximately 95% of the retrotransposons in the VAP-1 genome. These elements are known to enhance virulence during plant infection, and their derepression, followed by subsequent silencing, can lead to a reduction in virulence (Fouché et al., 2020).
ATP-driven efflux pumps, members of the ATP-binding cassette (ABC) superfamily, are critical for the transport of substrates across cellular membranes. Phytopathogenic fungi encounter toxic environments during infection as a result of plant defense responses. ABC transporters are essential for host infection, as they enable fungi to protect themselves against plant defense mechanisms (Urban et al., 1999). Gene Ontology (GO) annotations of proteins in the VAP-1 genome reveal that “ATP binding” is the most significantly enriched term in the “molecular function” domain. Within the “cellular component” domain, “membrane” is the most enriched category for VAP-1 proteins. In pathogenic filamentous fungi, the curvature of the cytoplasmic cell membrane is a contributing factor to fungal virulence (Peng and Chen, 2023). Furthermore, phytopathogens release toxins to facilitate host entry, targeting the plant cell membrane, mitochondria, and chloroplasts, thereby disrupting plant cellular integrity or metabolism (Shang et al., 2016). Virulence factors in pathogenic fungi, such as phospholipases, lipases, proteases, hemolysins, phosphatases, ureases, and other hydrolases, form an effective arsenal for successful penetration, survival, and dissemination within the host (Satala et al., 2023). GO annotation of VAP-1 proteins highlights “proteolysis” as the most significantly enriched term in the “biological process” domain.
Comparative genomics analysis of the VAP-1 genome with R. solani genomes infecting rice, sugar beet, and tobacco reveals a closer species-level relationship between VAP-1 and the rice isolate, with a greater degree of genomic divergence observed in the isolates infecting sugar beet and tobacco. It has been demonstrated that closely related species often exhibit shared infection strategies and virulence mechanisms, including the production of mycotoxins and tissue-degrading enzymes (Aoki and O’Donnell, 1999). As pathogenic fungi adapt to different host plants and environmental conditions, they can evolve distinct pathogenic strategies. Evolutionary divergence among species can lead to variations in virulence and infection mechanisms (Talbot, 2003). Orthologous clustering of VAP-1 protein sequences with those from R. solani infecting rice, sugar beet, and tobacco resulted in a single large cluster comprising 4,028 proteins common to all fungal genomes, while a distinct singleton cluster of 105 proteins was unique to the VAP-1 genome. Gene Ontology (GO) analysis of these 105 VAP-1 specific proteins revealed significant enrichment in pathways related to oxalate metabolic processes, pathogenesis, polysaccharide catabolic processes, and carbohydrate catabolic processes. The oxalate metabolic process is critical for the pathogenicity of plant pathogenic fungi, contributing to infection, colonization, and tissue damage by altering the host plant’s pH, degrading cell walls, disrupting calcium signaling, and suppressing immune responses (Dutton and Evans, 1996). R. solani utilizes various pathogenic factors, including secondary metabolites, carbohydrate-active enzymes, secreted proteins, and effectors to facilitate infection and virulence against its host plants (Li et al., 2021). During infection, R. solani secretes a range of cell wall-degrading enzymes that break down complex polysaccharides in the plant cell wall, enabling the fungus to access the plant’s cellular contents and facilitate penetration and colonization of plant tissues (Xue et al., 2018). The degradation of plant cell wall polysaccharides through carbohydrate catabolism allows R. solani to adapt to different plant environments by utilizing various plant-derived carbohydrates (Majumdar et al., 2022). This metabolic flexibility in nutrient acquisition enhances the pathogen’s ability to infect a wide range of host plants, including rice, sugar beet, tobacco, and, as shown in our study, proso millet. Comparative genomics has significantly advanced our understanding of fungal pathogens, offering critical insights into their genetic composition, virulence factors, and mechanisms of adaptation. This knowledge is pivotal for the development of innovative disease management strategies, such as the design of targeted fungicides, biocontrol agents, and the breeding of resistant plant varieties. Despite challenges, particularly those related to genomic complexity and the evolution of resistance, comparative genomics remains an invaluable tool for addressing fungal diseases in enhancing plant health.
Secretory proteins encoded in the VAP-1 genome are classified into several functional categories, including MFS general substrate transporters, (Trans) glycosidases, P-loop containing nucleoside triphosphate hydrolases, and galactose oxidase within the central domain superfamily. The protein repertoire, particularly carbohydrate-active enzymes (CAZymes), play a pivotal role in degrading plant cell wall polymers and releasing cell wall-bound saccharides, which serve as carbon sources for the actively infecting fungus (Hage and Rosso, 2021). Among the CAZymes identified in the VAP-1 genome, glycoside hydrolases constitute the largest group, followed by auxiliary activities, polysaccharide lyases, glycosyltransferases, and carbohydrate esterases. Notably, cellulases, xylanases, and pectinases, which degrade polysaccharides, are part of the glycoside hydrolase family and may play critical roles as pathogenicity factors in the infection of wheat spikes by Fusarium graminearum (Wanjiru et al., 2002). Glycoside hydrolases are particularly significant as they degrade plant cell wall polysaccharides, facilitating fungal penetration and colonization (Kubicek et al., 2014). Similarly, AAs contribute to oxidative degradation of lignocellulosic components, further assisting in tissue maceration (Hemsworth et al., 2015). Auxiliary activities represent a diverse group of enzymes that support glycoside hydrolases by catalyzing the oxidation of carbohydrates, often in synergy with other enzymatic activities. This collaboration enhances the degradation of plant cell walls, facilitating fungal invasion and colonization (Levasseur et al., 2013). Additionally, glycosyltransferases (GTs), enzymes responsible for catalyzing the transfer of sugar moieties to acceptor molecules, play a crucial role in fungal pathogenesis. For example, glycosyltransferases are involved in regulating fumonisin biosynthesis and virulence in Fusarium verticillioides, a major global pathogen of maize, highlighting their importance in the infection process (Deng et al., 2021). The ability of R. solani to produce a diverse array of CAZymes indicates its efficiency in degrading plant defenses and sustaining infection.
Rhizoctonia
Comments (0)